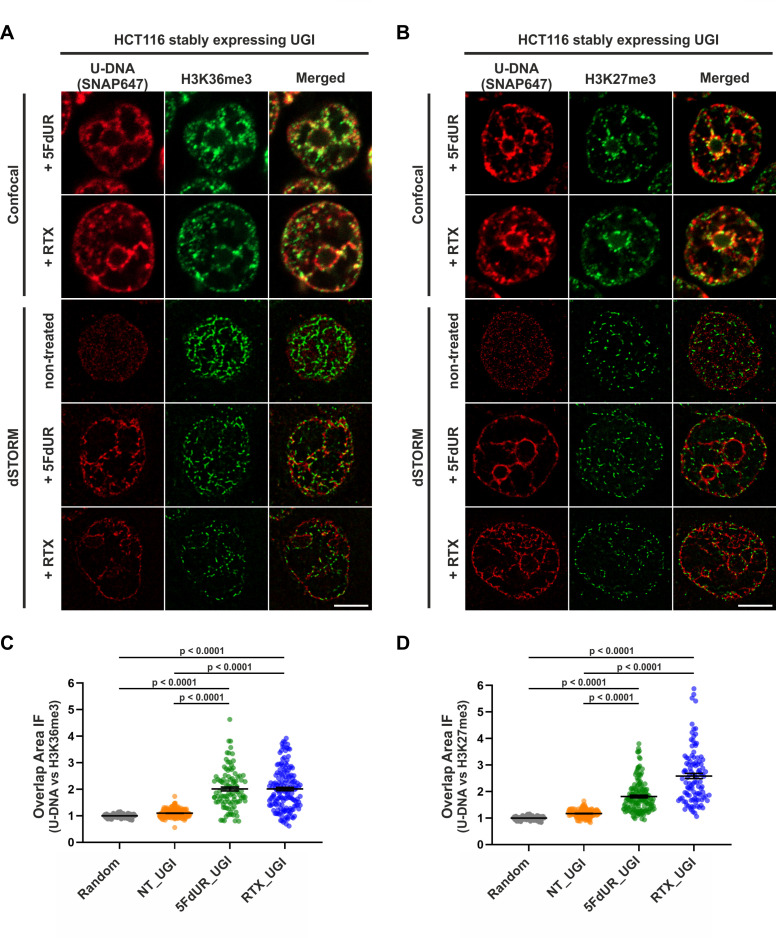
Figure 8. Genomic uracil moieties colocalize with H3K36me3 and H3K27me3 analyzed by super-resolution microscopy.
Confocal and dSTORM imaging were performed on non-treated, 5FdUR or RTX treated HCT116 cells stably expressing UGI to compare the localization of genomic uracil residues (red) to histone markers, H3K36me3 (green) (A) or H3K27me3 (green) (B). Scale bar represents 5 μm. The graphs display the cross-pair orrelation analysis between U-DNA and H3K36me3 (C) or H3K27me3 (D) performed on dSTORM images. Overlap is defined as any amount of pixel overlap between segmented objects. Total numbers of analyzed nuclei for H3K36me3 staining (C) were the following: NT_UGI (n = 205), 5FdUR_UGI (n = 101) and RTX_UGI (n = 153) from two independent experiments. Total numbers of analyzed nuclei for H3K27me3 staining (D) were the following: NT_UGI (n = 154), 5FdUR_UGI (n = 151) and RTX_UGI (n = 107) from two independent experiments. Black line denotes the mean of each dataset, and error bars represent standard errors of the mean (SEM). The color code follows the one in Figure 3A. Source data are available in Figure 8—source data 1.