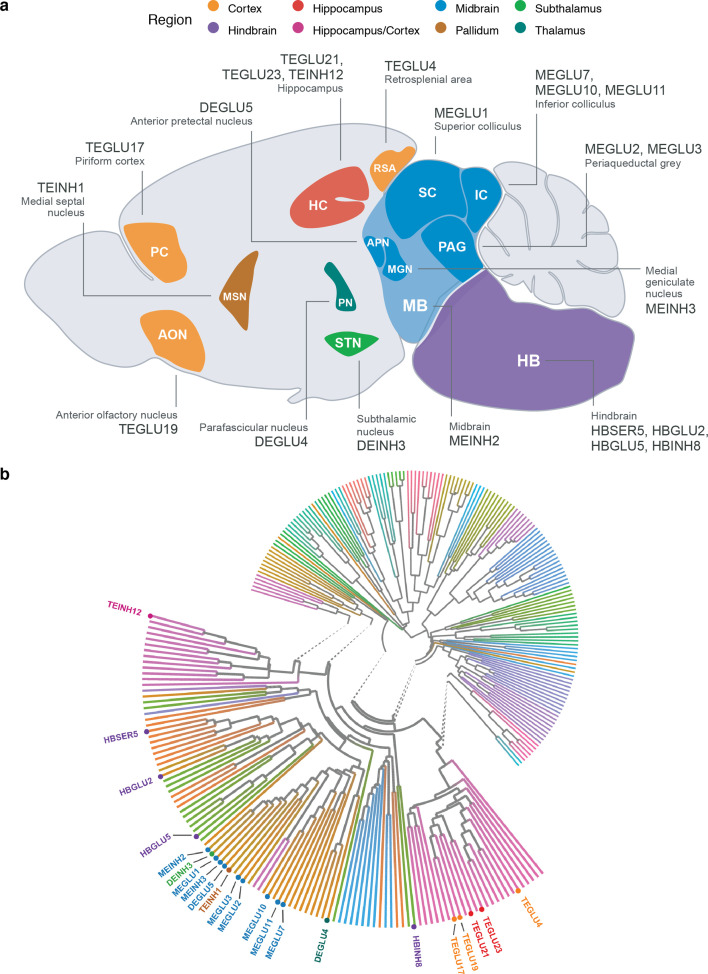
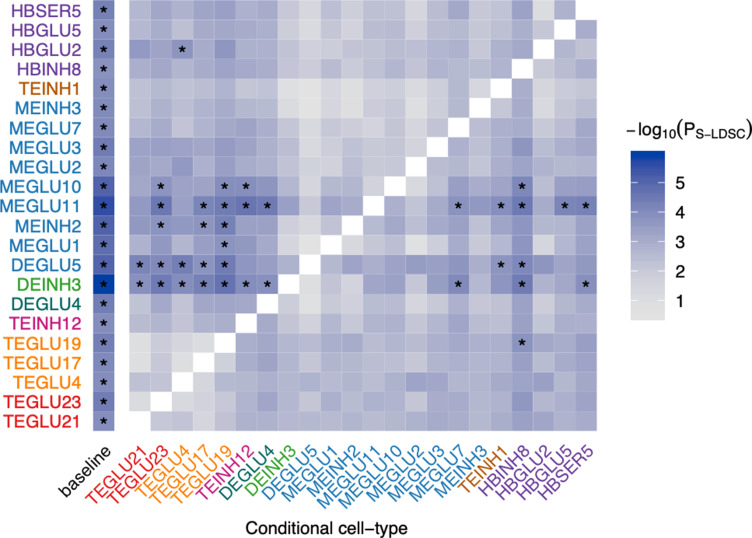
Figure 4. Neuroanatomical location and transcriptional similarity of brain cell types enriching for BMI GWAS variants.
(a) Sagittal mouse brain view showing the 22 BMI GWAS-enriched cell types. The first two letters in each cell type label denote the developmental compartment (ME, mesencephalon; DE, diencephalon; TE, telencephalon), letters three to five denote the neurotransmitter type (INH, inhibitory; GLU, glutamatergic) and the numerical suffix represents an arbitrary number assigned to the given cell type. (b) Circular dendrogram showing the similarity of all Mouse Nervous System dataset cell type expression specificity (ESμ) values. Dendrogram edges colored by taxonomy described in Zeisel et al., 2018. Expectedly, the cell types clustered according to their neuroanatomical origin. For clarity, only the labels of the 22 BMI GWAS enriched cell types are shown.