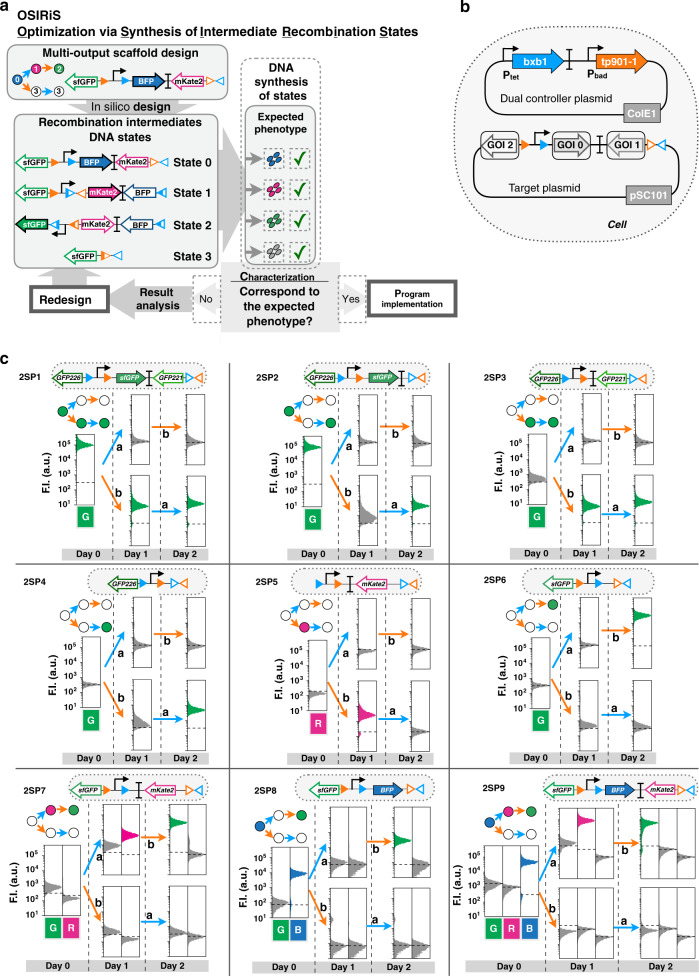
Fig. 2. Characterization of 2-input single-lineage programs.
a OSIRiS workflow. A 2-input history-dependent scaffold with consecutive expressions of BFP, RFP, and GFP in one lineage is designed. The DNA sequences of the different input states corresponding to the intermediate recombination states are generated (DNA states). The DNA states sequences are synthesized and characterized and the phenotypes are compared to the expected ones. If they match, the implementation of the program is performed. Otherwise, the results are precisely analyzed to identify the origin of the failure, the multioutput scaffold is redesigned and a new OSIRiS cycle is performed. b genetic design of the two plasmid used to characterize each program. The dual-controller plasmid regulates the gene expression of the Bxb1 and Tp901-1 integrases. The target plasmid corresponds to the 2-input history-dependent scaffold. c Nine single-lineage history-dependent programs (2SP1–2SP9) exhibiting lineage specific, or state-specific gene expression with single or multiple genetic outputs were implemented and characterized. Bxb1 and Tp901-1 are induced by aTc (input a) and by arabinose (input b), respectively. The lineage tree for each program and its corresponding genetic DNA device are represented. Cells transformed with both plasmids were sequentially induced twice for 16 h, at which point fluorescence intensity was measured by flow cytometry. Each histogram shows the expression of fluorescent reporters expressed at different induction states. All experiments were performed in triplicate three times on three different days. A representative example from three biological replicates is depicted here. Fold change measurements can be found in Fig. S5.