Figure 8.
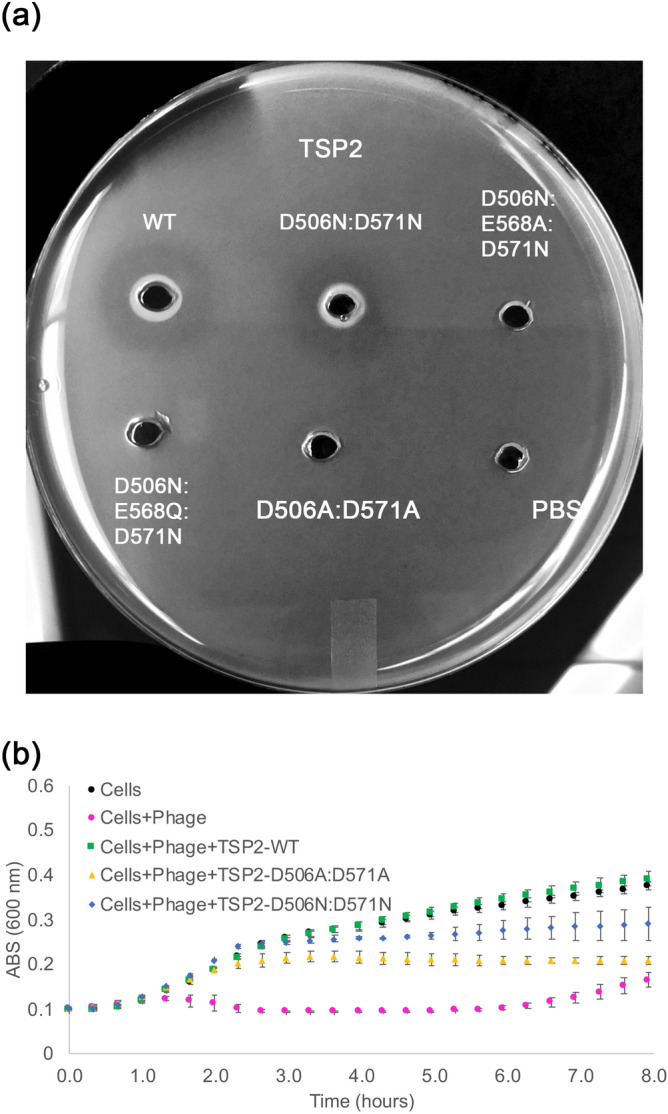
Assays to probe TSP2 active site mutants. Structure and mechanism-based analyses were used to identify the active site residues. (a) In the halo assay, E. coli strain ATCC 700,728 was embedded in agarose. Wells (3 mm) were cut out of the agarose and loaded with 10 μL (6 mg/mL) of wild-type TSP2 or active site TSP2 mutants, and incubated overnight at 37 °C to visualize glycosidase activity. The D506A:D571A TSP2 mutant was incapable of producing a halo, indicating glycosidase activity of TSP2 was abolished. Conversely, the D506:D571N TSP2 mutant retained enzymatic activity. However, introducing the E568A (i.e. D506N:E568A:D571N) or E568Q (i.e. D506N:D568Q:D571N) mutations to a D506N:D571N background inhibited TSP2 glycosidase activity, suggesting involvement of the Glu568 carboxyl group as part of an alternative catalytic machinery. Wild-type TSP2 and PBS only served as positive and negative controls for glycosidase activity, respectively. (b) In the bacterial infection assay, E. coli O157:H7 (ATCC 700728) culture was mixed with TSP variants as described in Methods. Absorbance measurements at 600 nm were made every 20 min. Bacterial growth is shown for E. coli incubated with phage following treatment with 100 μg/mL of either wild-type or TSP2 (green squares, D506:D571A TSP2 (yellow triangles), or D506N:D571N TSP2 (blue diamonds). Growth of bacterial cells alone (black circles) and non-TSP treated bacterial cells incubated with phage (magenta circles) served as controls. The mean values of technical triplicate and the standard deviations are shown.
