Abstract
Cloned animals generated by somatic cell nuclear transfer (SCNT) have been reported for many years; however, SCNT is extremely inefficient, and zygotic genome activation (ZGA) is required for SCNT‐mediated somatic cell reprogramming. To identify candidate factors that facilitate ZGA in SCNT‐mediated reprogramming, we performed siRNA‐repressor and mRNA‐inducer screenings, which reveal Dux, Dppa2, and Dppa4 as key factors enhancing ZGA in SCNT. We show that direct injection of ZGA inducers has no significant effect on SCNT blastocyst formation; however, following the establishment of an inducible Dux transgenic mouse model, we demonstrate that transient overexpression of Dux not only improves SCNT efficiency but also increases that of chemically induced pluripotent stem cell reprogramming. Moreover, transcriptome profiling reveals that Dux‐treated SCNT embryos are similar to fertilized embryos. Furthermore, transient overexpression of Dux combined with inactivation of DNA methyltransferases (Dnmts) further promotes the full embryonic development of SCNT‐derived animals. These findings enhance our understanding of ZGA‐regulator function in somatic reprogramming.
Keywords: chemically induced pluripotent stem cells, Dppa2/4, Dux, SCNT, zygotic genome activation
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Development & Differentiation; Regenerative Medicine
Transient overexpression of ZGA‐inducer Dux not only enhances the efficiency of SCNT, but also of chemically‐induced somatic cell reprogramming. Combined with inactivation of DNA‐methyltransferases, it further improves the full‐term development of cloned embryos.
Introduction
Following successful cloning of the first sheep (Wilmut et al, 1997), at least 20 other mammalian species have been cloned (Rodriguez‐Osorio et al, 2012), including non‐human primate monkeys (Liu et al, 2018). Indeed, successful cloning of these animals made reproductive cloning possible and introduced the possibility of therapeutic cloning (Rideout et al, 2002; Hochedlinger & Jaenisch, 2003). Somatic cell nuclear transfer (SCNT) is the technical term used to describe a process in which a terminally differentiated somatic cell is transferred into an enucleated oocyte, followed by reprogramming of the nucleus by the oocyte cytoplasmic factors to attain a zygote‐like state (Matoba & Zhang, 2018). Both SCNT‐derived and “Yamanaka factor”‐induced pluripotent stem (iPS) cells have the potential to generate patient‐specific pluripotent stem cells for replacement therapy; however, compared with iPS reprogramming, SCNT reprogramming represents an oncogene‐free methodology. Therefore, this makes SCNT‐derived pluripotent stem cells more suitable for human therapeutic applications.
Following fertilization, the newly formed zygotic genome is activated through a process known as zygotic genome activation (ZGA), which promotes subsequent zygote development into an adult animal (Lu & Zhang, 2015). A similar mechanism is likely used in ZGA of SCNT embryos (Gao et al, 2007; Matoba & Zhang, 2018). Indeed, our and other previous studies revealed incomplete ZGA as a major reprogramming barrier (Matoba et al, 2014; Liu et al, 2016; Wang et al, 2018; Yang et al, 2018). Although many advances have been made in SCNT technology via different epigenetic regulators, SCNT for producing cloned animals remains inefficient.
The transcription factor of double homeobox (Dux) was identified as a key inducer of ZGA in normal fertilized embryos (De Iaco et al, 2017; Hendrickson et al, 2017; Whiddon et al, 2017). Recently, two independent studies found that the transcription factors, developmental pluripotency‐associated 2 (Dppa2) and developmental pluripotency‐associated 4 (Dppa4), are both necessary and sufficient for the activation of ZGA‐related genes in embryonic stem cells (ESCs) (De Iaco et al, 2019; Eckersley‐Maslin et al, 2019); however, aside from Dux, Dppa2/4 as in vivo inducer that activate ZGA transcripts remain elusive. Therefore, as far as we know, at least three factors can be called master ZGA inducer in ESCs. Furthermore, Dux overexpression is sufficient to drive pluripotent stem cells into a totipotent state by promoting the expression of two‐cell embryo‐specific transcripts (known as 2‐cell‐like stem cells) (Hendrickson et al, 2017; Fu et al, 2019; Yang et al, 2020). Additionally, several studies report that a totipotent state can be achieved by depleting ZGA repressor, such as tripartite motif‐containing 28 (Trim28; also known as KRAB‐associated protein 1, Kap1) (Rowe et al, 2010), microRNA 34a (miR‐34a) (Choi et al, 2017), chromatin assembly factor (CAF‐1) (Ishiuchi et al, 2015), lysine (K)‐specific demethylase 1A (Lsd1/Kdm1a) (Macfarlan et al, 2012; Ancelin et al, 2016; Wasson et al, 2016), and long interspersed nuclear element (LINE1) (Percharde et al, 2018).
The effect of ZGA regulators in somatic cell reprogramming remains unknown. Thus, this study aimed to explore the relationship between ZGA regulators and SCNT efficiency and to identify a smart factor that could promote the efficiency of somatic reprogramming.
Results and Discussion
We have recently established a MERVL::tdTomato‐based real‐time monitoring system for ZGA (Yang et al, 2018). To identify candidate factors that facilitate ZGA in SCNT reprogramming, we performed small‐interfering (si)RNA‐repressor and mRNA‐inducer screening in SCNT (Fig 1A). From the screen, we identified a number of factors that when overexpression or knockdown resulted in reporter activation (Appendix Fig S1A). Among the identified factors, Dux, Dppa2, and Dppa4 showed the strongest phenotype (Fig 1B and C). Interestingly, we found that most SCNT embryos injected with Dppa2 or Dppa4 mRNA arrested at the 1‐cell stage (called “1‐cell block” for simplicity; Fig 1D and E; [Link], [Link]), whereas direct injection of Dux mRNA showed no significant effect on the blastocyst formation rate (vs. canonical SCNT; 21.2 vs. 23.6%; Fig 1D; Appendix Fig S1B and Table S1). Indeed, direct injection of Dux will increase the rate of SCNT embryo fragmentation (Fig 1D and E; Movie EV2). Nevertheless, when Dux was over‐expressed in SCNT embryos, the rate of 2‐cell block was significantly reduced (from 54.7 to 9.6%; Fig 1D; Appendix Table S1). These results suggested Dux (but not Dppa2/4) as a key factor whose overexpression can rescue 2‐cell arrest in SCNT embryos.
Figure 1. Dux promotes SCNT‐mediated somatic cell reprogramming.
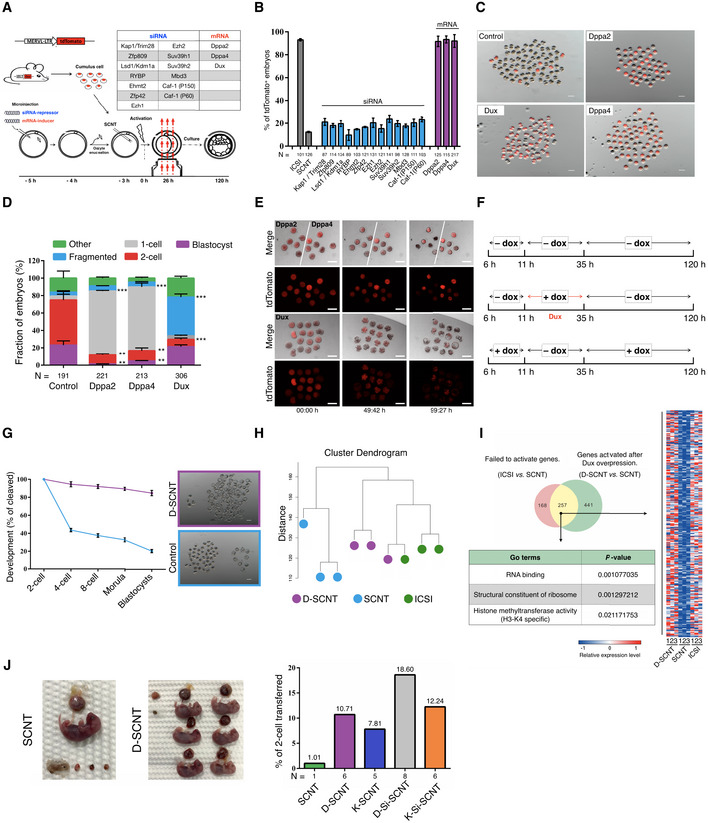
- Schematic illustration of the screening strategy.
- Quantification of embryos that expressing the ZGA reporter (MERVL::tdTomato) after injection with a siRNA‐repressor or mRNA‐inducer. N, the total number of embryos analyzed for each condition. Error bars, mean ± SD; n = 3 biological replicates per group.
- Representative fluorescence image of SCNT embryos derived by different methods. The SCNT embryos produced by transfer of MERVL::tdTomato cumulus cells (B6D2F1 background) into WT enucleated oocytes. n = 3 biological replicates per group. Scale bar, 100 μm.
- Stacked bar plots show the fraction of embryos at blastocyst stages after injection with different mRNA, as indicated. Error bars, mean ± SD; n = 3 biological replicates per group; N, total number of embryos analyzed for each condition; **P < 0.01, ***P < 0.001 as compared with control group, by two‐tailed Student's t‐test.
- Representative live‐cell images of dynamic ZGA‐reporter expression during SCNT embryo development. The time after initial observation is shown at the bottom of the image. n = 3 biological replicates per group. Scale bar, 50 μm.
- Schematic transient induction of Dux expression. All timings reported in this paper are hour post‐activation.
- Preimplantation development of SCNT embryos. The percentage of embryos reaching each indicated stage is shown. The D‐SCNT embryos produced by transfer of dox‐Dux cumulus cell (B6D2F1 background) into WT enucleated oocytes (B6D2F1 background). Error bars, mean ± SD; n = 3 biological replicates per group. Scale bar, 100 μm.
- Unsupervised hierarchical clustering.
- Venn diagram showing the overlap between the genes that failed to be activated in SCNT 2‐cell embryos and derepressed in D‐SCNT. Heat map, GO terms showing the expression pattern of 257 overlapping genes.
- Image of full‐term cloned mice derived by canonical SCNT and D‐SCNT method (left). The birth rate of SCNT embryos derived by different methods is indicated (right). N, the total number of cloned pups obtained from each condition.
Zygotic genome activation is governed by a time‐dependent “zygotic clock” (i.e., 24‐h post‐fertilization), and Dux is expressed as a brief pulse in early 2‐cell stage (Wiekowski et al, 1991; Schultz, 1993; Nothias et al, 1995, 1996; Eckersley‐Maslin et al, 2018). Therefore, we wonder whether Dux is time‐dependent on improving the efficiency of SCNT embryo development. To answer this question, we generated transgenic mouse lines containing a doxycycline (dox)‐inducible Dux gene (Appendix Fig S2A and B). We first confirmed that the dox‐inducible system did not influence developmental capacity (Appendix Fig S2C and D, Table S2). By switching between dox‐containing and dox‐free culture medium at various time points, we mapped the critical period for dox‐Dux activity to a 24‐h window (Fig 1F). When the dox was supplied in this window, the SCNT blastocyst formation rate was significantly increased (Fig 1G; Appendix Table S2). The induction efficiency of Dux upon dox treatment was confirmed by single‐embryo quantitative reverse transcription PCR (RT‐qPCR; Fig EV1A). Moreover, single‐embryo RT‐qPCR and immunofluorescence (IF) results also showed that overexpression of Dux could significantly increase the expression of ZGA‐related Zscan4 and MERVL genes (Fig EV1A and B). We subsequently performed single‐cell RNA sequencing (scRNA‐seq) to examine possible changes in ZGA‐related gene expressions in Dux‐overexpressing 2‐cell SCNT embryos (D‐SCNT). Unsupervised hierarchal clustering revealed that the intracytoplasmic sperm injection (ICSI) and D‐SCNT generated embryos were in the same cluster and that levels of ZGA‐related genes were upregulated in D‐SCNT‐generated embryos (Fig 1H and I; Appendix Fig S3A–D; [Link], [Link]). These results indicating the improvement of Dux on SCNT embryonic development were dependent on its treatment time.
Figure EV1. Dux is required for SCNT‐mediated somatic cell reprogramming.
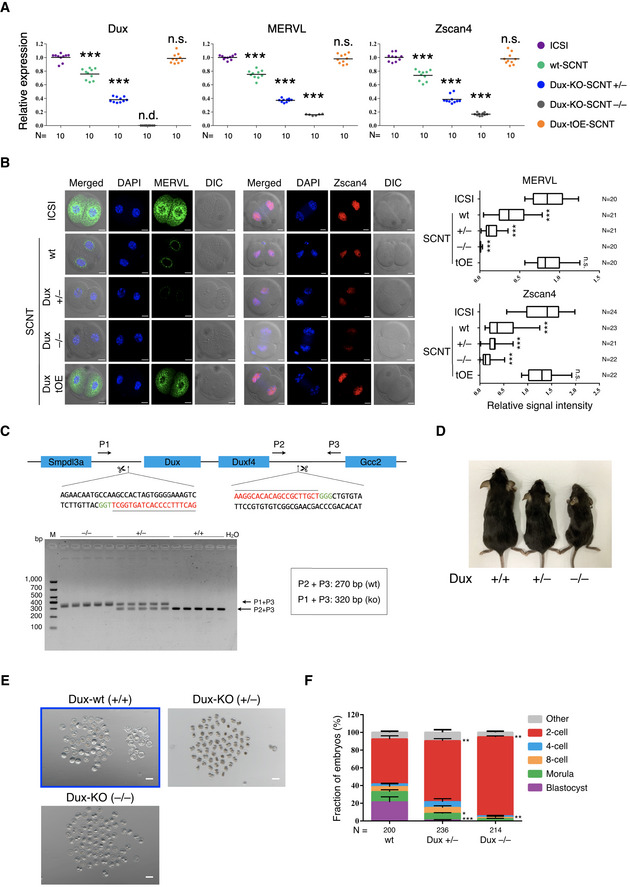
- Single‐embryo RT‐qPCR analysis of Dux and select ZGA‐related genes in 2‐cell embryos derived from ICSI, wt‐SCNT, Dux‐KO-SCNT, and Dux‐tOE-SCNT. Data shown are expression values relative to glyceraldehyde‐3-phosphate dehydrogenase (GAPDH); the value in ICSI group was set as 1; each dot represents one embryo; black bars indicate mean value; KO, knockout; tOE, transient overexpression; wt, wild‐type; +/−, mono‐allelic deletion; −/−, bi‐allelic deletion; N, total number of embryos analyzed for each condition; ***P < 0.001 as compared with the ICSI group, by two‐tailed Student's t‐test. n.s., not significant. n.d., not detectable.
- Immunofluorescence images of ICSI, wt‐SCNT, Dux‐KO-SCNT, and Dux‐tOE-SCNT embryos (left). Scale bar, 20 μm. The box plots for relative intensities of different antibodies from n = 3 independent experiments (right). The middle line in the box indicates the median, the edges indicate the 25th/75th percentiles, and the whiskers indicate the 2.5th/97.5th percentiles; KO, knockout; tOE, transient overexpression; wt, wild‐type; +/−, mono‐allelic deletion; −/−, bi‐allelic deletion; N, total number of embryos analyzed for each condition; ***P < 0.001 as compared with the ICSI group, by two‐tailed Student's t‐test. n.s., not significant.
- Schematic view of the Dux‐KO strategy and the PCR results of the genomic DNA. Arrows indicate primer locations. The sgRNA sequence and CRISPR proto‐spacer adjacent motif (PAM) sequence are indicated in red and green, respectively. P, primer; M, marker.
- Dux‐KO mice were frequently smaller than their age‐matched wild‐type littermates. wt, wild‐type; +/−, mono‐allelic deletion; −/−, bi‐allelic deletion.
- Images of SCNT embryos derived from wild‐type and Dux‐KO cumulus cells after 120 h of culturing in vitro. Images are representative examples of the quantification shown in Fig EV1F. The images in the blue rectangle are the same in Appendix Fig S2C. wt, wild‐type; +/−, mono‐allelic deletion; −/−, bi‐allelic deletion. Scale bar, 100 μm.
- Stacked bar plots showing the fraction of SCNT embryos at blastocyst stages. N, the total number of embryos analyzed for each condition. Error bars, mean ± SD; n = 3 biological replicates per group; *P < 0.05, **P < 0.01, ***P < 0.001 according to two‐tailed Student's t‐test.
To further determine whether Dux is a key regulator of ZGA in SCNT reprogramming, we generated Dux‐knockout (Dux‐KO) mouse lines using CRISPR/Cas9 (Fig EV1C and D), as previously described (Fu et al, 2019), and used somatic cells from Dux‐KO mice as nuclear donors for SCNT. We found that only 1.4% Dux+/− heterozygous SCNT embryos developed to the blastocyst stage, whereas none of the Dux−/− homozygous SCNT embryos reached the blastocyst stage (Fig EV1E and F; Appendix Table S3). Notably, most of Dux−/− SCNT embryos were blocked at the 2‐cell stage (94.1% in Dux−/−‐SCNT, 75.3% in Dux+/− ‐SCNT, and 55.6% in wild‐type [wt]‐SCNT). This was a more severe phenotype than that reported in recent studies in fertilized Dux‐KO embryos (Chen & Zhang, 2019; Guo et al, 2019), suggesting that Dux is a critical factor for SCNT reprogramming. Furthermore, single‐embryo RT‐qPCR results showed that the expression of Dux and ZGA‐related genes was significantly decreased in Dux−/−‐KO SCNT embryos relative to Dux+/−‐KO‐ and WT‐SCNT embryos (Fig EV1A). As no commercial antibody for Dux is available, we verified the single‐embryo qRT‐PCR results by IF in ZGA‐related genes (Fig EV1B).
The SCNT blastocyst rate was improved by injecting Kdm4d mRNA (K‐SCNT) (Matoba et al, 2014). We then compared the developmental potential of SCNT embryos derived by K‐SCNT and D‐SCNT, finding that the blastocyst formation rate of D‐SCNT was similar to that of K‐SCNT (84.72 vs. 86.79%; Appendix Fig S4A, Table S2). Nonetheless, the cloned pup‐birth rate of D‐SCNT embryos was higher than that of K‐SCNT embryos (10.71 vs. 7.81%; Fig 1J; Appendix Table S4). Despite D‐SCNT or K‐SCNT, the abnormally large placentas are still observed in those cloned pups (Appendix Fig S4B, Table S4). Previous studies indicated that knockdown of DNA methyltransferases Dnmt3a/b led to less placental abnormalities (Si‐SCNT) (Gao et al, 2018). Therefore, we investigated whether combined D‐SCNT and Si‐SCNT could further improve SCNT embryonic development. Compared with canonical SCNT, combined K‐SCNT and Si‐SCNT improved the pup‐birth rate from 1.01 to 12.24%, but less than that observed for D‐SCNT combined with Si‐SCNT (18.60%; Fig 1J; Appendix Table S4). Notably, the large placental phenotype was rescued by the D‐Si‐SCNT combined approach (Appendix Fig S4B, Table S4). Genotyping analysis confirmed that all of the cloned pups were generated from donor cells (Appendix Fig S4C).
Recently, Zhao et al identified a unique embryonic 2‐cell‐like status as the key molecular event during chemically induced pluripotent stem cells (CiPSCs) induction (Fig 2A) (Zhao et al, 2018); therefore, we attempted to improve chemical induction by Dux overexpression. The dox‐inducible Dux mouse embryonic fibroblasts (dDux‐MEFs) were isolated from the transgenic mice. As shown in Fig 2B, we treated the dDux‐MEFs with dox for various durations and observed that Dux displayed remarkable effect at the stage‐2 of chemical induction (hereinafter referred to as D‐CiPS). We observed ~ 36 alkaline phosphatase‐positive (AP+) colonies in wells undergoing D‐CiPS induction (starting from 50,000 MEFs per well of six‐well plate), whereas only 26 AP+ colonies were observed in wells undergoing canonical CiPS induction (Fig 2C and D). To further test the pluripotency of iPS cells, we established stable D‐CiPSCs lines using the D‐CiPS method. Interestingly, we found that levels of ZGA‐related genes were upregulated in dox‐untreated D‐CiPSCs relative to those generated by canonical chemical induction at passage one (Fig 2E). We confirmed that these D‐CiPSCs were pluripotent according to standard characterization procedures (Fig 2F–I), and bisulfite sequencing revealed that successful demethylation in the promoters of Oct4 and Nanog (Fig 2J).
Figure 2. Dux facilitates chemical‐mediated somatic cell reprogramming.
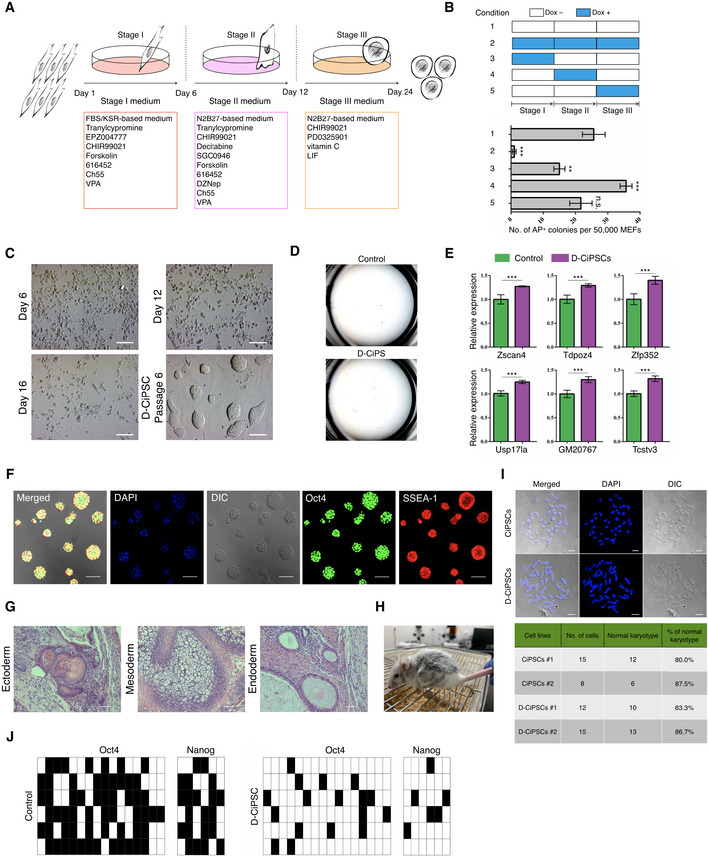
- Schematic diagram for the induction of CiPSCs from MEFs using the “three‐step” method (Zhao et al, 2018).
- Durations of dox addition at different time points during chemical induction (upper) and assessment of AP+ colonies (bottom). Error bars, mean ± SD; n = 3 biological replicates per condition; **P < 0.01, ***P < 0.001 as compared with condition‐1, by two‐tailed Student's t‐test. n.s., not significant.
- Morphological changes at distinct time points during D‐CiPS induction. Scale bar, 100 μm.
- Representative images of the number of AP+ positive colonies. n = 5 biological replicates per condition.
- RT‐qPCR analysis of select ZGA‐related genes in dox‐untreated D‐CiPSCs and canonical CiPSCs at passage 1. The value in CiPSCs was set as 1, and data shown are mean expression values relative to GAPDH. Error bars, mean ± SEM; n = 3 biological replicates per group; ***P < 0.001 by two‐tailed Student's t‐test.
- Representative immunostaining images of D‐CiPSCs expressing pluripotency markers. n = 5 biological replicates. Scale bar, 50 μm.
- Histology by H&E staining of teratoma tissues derived by D‐CiPS cells. n = 3 biological replicates. Scale bar, 200 μm.
- Chimeric mouse generated from D‐CiPSCs.
- Karyotype analysis of D‐CiPSCs and canonical CiPSCs. The majority of D‐CiPSCs and CiPSCs cells possessed a normal karyotype. Scale bar, 10 μm.
- Bisulfite sequencing analysis of demethylation of Oct4 and Nanog promoters in D‐CiPSCs or canonical CiPSCs. Filled and empty squares represent methylated and unmethylated CpGs, respectively.
RNA‐seq analysis confirmed that Dux promoted the expression of ZGA‐related genes during chemical induction at stage 2 (Fig EV2A–C; Appendix Fig S5A–C). Notably, most of MERVL long terminal repeat (LTR)‐driven genes were also upregulated by Dux overexpression as compared with canonical chemical induction at stage 2 (Fig EV3D; Appendix Fig S5D). Additionally, the RNA‐seq results showed that Dux overexpression increased the expression of genes related to pluripotency (Fig EV3E). Among these upregulated ZGA‐related genes, Zscan4 and Tcstv1, knockdown of which impaired CiPS cell induction, participate with Dppa family members and Oct4 to mediate pluripotency transition and telomere elongation (Xu et al, 2015; Fu et al, 2018; Zhao et al, 2018; Yang et al, 2019). Moreover, human DUX4, as a conserved homolog of mouse Dux, recruits the histone acetyltransferases p300/CBP to drive epigenetic changes (Choi et al, 2016; Bosnakovski et al, 2019). These results might explain why Dux improved chemical induction; however, further studies are required to evaluate the ability of Dux to modulate telomerase activity and DNA/histone modifications in CiPSCs induction.
Figure EV2. Dux improves chemical induction via activated expression of ZGA‐related genes.
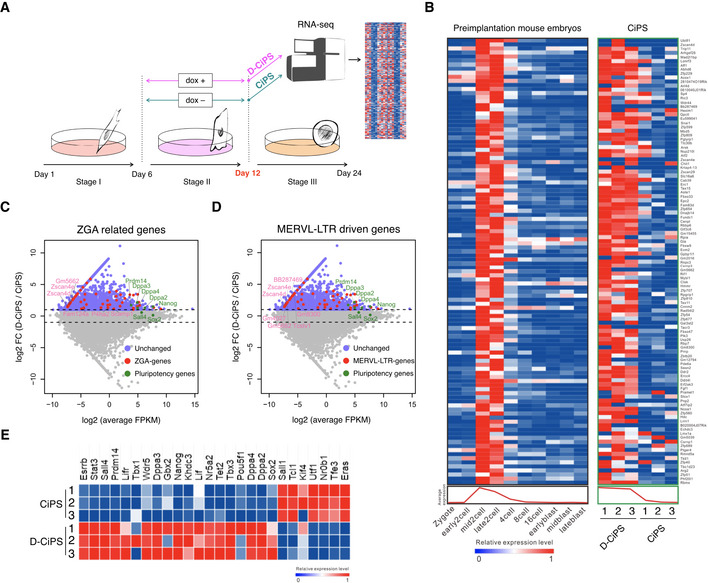
-
ASchematic illustration of the RNA‐seq strategy.
- B
-
C, DMA plots showing differentially expressed ZGA‐related genes (C) and MERVL‐LTR-driven genes (D) during D‐CiPS induction. n = 3 RNA‐seq replicates per condition. Upregulated genes are in violet; MERVL‐LTR-driven and ZGA‐related genes are in magenta; key genes are annotated.
-
EHeat map comparing the expression of master pluripotent factors between D‐CiPS and canonical CiPS induction. n = 3 RNA‐seq replicates per condition.
Figure EV3. Dux is an activator but has cytotoxicity for the CiPSCs to 2C‐like cell transition.
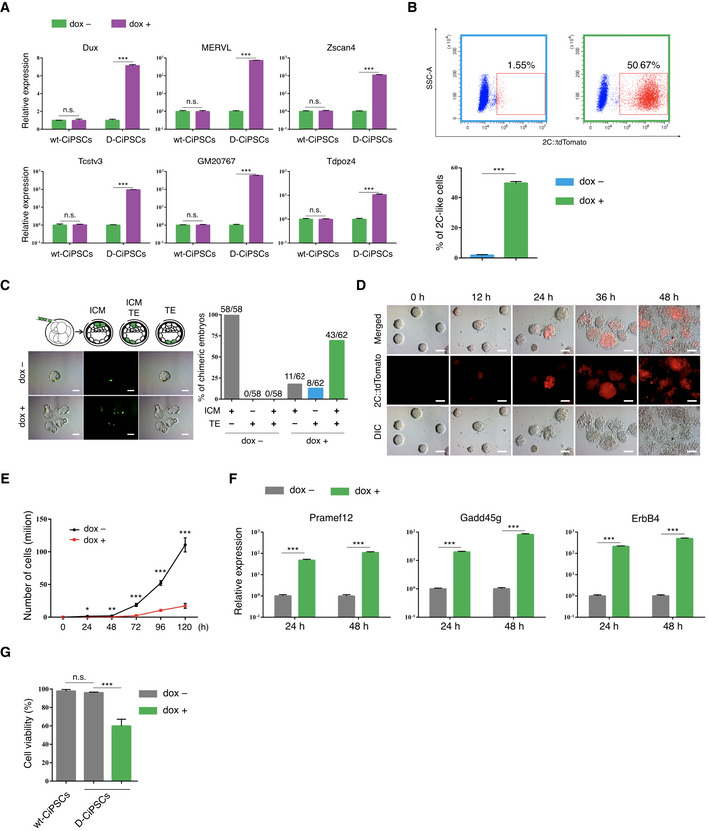
- RT‐qPCR analysis of select ZGA‐related genes in dox‐treated (24 h) and dox‐untreated D‐CiPSCs. The value in untreated D‐CiPSCs was set as 1, and data shown are mean expression values relative to GAPDH. Error bars, mean ± SEM; wt, wild‐type; n = 3 biological replicates per condition; ***P < 0.001 by two‐tailed Student's t‐test.
- Fluorescence‐activated cell sorting (FACS) analysis of 2C‐like cells in dox‐treated and dox‐untreated D‐CiPSCs. Error bars, mean ± SEM; n = 3 biological replicates per condition; ***P < 0.001 by two‐tailed Student's t‐test.
- Schematic illustration of the experimental approach (left up). Four enhanced green fluorescent protein (EGFP)‐labeled, dox‐treated, or dox‐untreated D‐CiPSCs were microinjected into each 8‐cell stage embryo, and the chimeric embryo was in vitro cultured until the blastocyst stage. Representative fluorescence images of injected GFP‐labeled dox‐treated or dox‐untreated D‐CiPSCs differentiating to the inner cell mass (ICM) and trophectoderm (TE) in blastocysts (left bottom). The percentage of chimeric blastocysts with D‐CiPSCs contribution to ICM, TE, and ICM + TE (right). n = 3 biological replicates per group. N, the total number of embryos analyzed for each condition. Scale bar, 20 μm.
- Representative images of D‐CiPSCs at 12‐, 24-, 36‐, and 48‐h post‐dox induction. n ≥ 3 biological replicates per condition. Scale bar, 50 μm.
- Cell proliferation of D‐CiPSCs following treatment with or without dox. Error bars, mean ± SEM; n = 3 biological replicates per condition; *P < 0.05, **P < 0.01, ***P < 0.001 by two‐tailed Student's t‐test.
- RT‐qPCR analysis of Dux induced expression of apoptosis‐related genes in dox‐treated (24 h) and dox‐untreated D‐CiPSCs. The value in untreated D‐CiPSCs was set as 1, data shown are mean expression values relative to GAPDH. Error bars, mean ± SEM; n = 3 biological replicates per condition; ***P < 0.001 by two‐tailed Student's t‐test.
- MTT cell viability assay confirmed that Dux influences the viability of D‐CiPSCs. Error bars, mean ± SD; wt, wild‐type; n = 3 biological replicates per condition; ***P < 0.001 by two‐tailed Student's t‐test.
Unlike pluripotent ESCs, totipotent ESCs (known as 2‐cell‐like cells, 2C‐like cells) express high levels of ZGA‐related genes, including MERVL and Zscan4, and are characterized by the capacity to contribute to both embryo and extraembryonic tissues (Macfarlan et al, 2012). As Dux can convert ESCs to 2C‐like cells (Hendrickson et al, 2017; Fu et al, 2019; Yang et al, 2020), we speculated that Dux could also convert CiPSCs to the 2C‐like state. To test this hypothesis, we labeled D‐CiPSCs by MERVL::tdTomato vector. As expected, dox treatment induced the expression of Dux and ZGA‐related genes in D‐CiPSCs (Fig EV3A), with fluorescence‐activated cell sorting (FACS) analysis revealing that Dux overexpression significantly increased the fraction of 2C‐like cells (Fig EV3B; Appendix Fig S5E). Furthermore, chimera‐formation assay showed that dox‐treated D‐CiPS cells exhibited a high incorporation frequency (43/62) into both the inner cell mass (ICM) and trophectoderm (TE), whereas untreated D‐CiPS cells were incapable (0/58) of TE incorporation (Fig EV3C). Notably, we observed rapid morphological changes, as D‐CiPSCs could not maintain the dome‐colony morphology upon Dux‐mediated induction in the first 36 h (Fig EV3D).
Previous reports indicate that human DUX4 and mouse Dux are toxic to C2C12 myoblasts, 3T3 fibroblasts, and ESCs (Bosnakovski et al, 2008, 2009). Consistently, we found that Dux overexpression not only decreased D‐CiPSC proliferation but also increased levels of apoptosis‐related genes induced by Dux in C2C12 cells (Eidahl et al, 2016) (Fig EV3E and F). Moreover, cell viability assays revealed that dox‐treated D‐CiPSCs consistently displayed significant decreases in cell viability (40%) relative to untreated D‐CiPSCs and WT ESCs (Fig EV3G). A recent study of Facioscapulohumeral muscular dystrophy found that the small‐molecule iP300w protects cells from DUX4‐mediated cytotoxicity (Bosnakovski et al, 2019), so it is possible that Dux overexpression combined with iP300w treatment allows the 2C‐like transition to run more smoothly by protecting cell viability. However, iP300w might influence totipotency by decreasing the expression of ZGA‐related genes, such as Zscan4 (Bosnakovski et al, 2019). Nevertheless, the regulatory relationship between Dux‐related totipotency and cytotoxicity regulated warrants further investigation.
When we prepare this manuscript, two recent studies suggested that Dux is important but not essential for fertilized‐embryo development via enhancing rather than ZGA inducer (Chen & Zhang, 2019; Guo et al, 2019). However, in the present study, we demonstrated that transient overexpression of Dux not only improved SCNT efficiency but also increased the efficiency of chemical reprogramming. Moreover, ectopic expression of Dppa2/4 in SCNT embryos resulted in 1‐cell block, implying that successful and timely ZGA is essential for the development of SCNT‐derived embryos. A recent study found that Dppa2/4 interacted with small ubiquitin‐like modifier‐related proteins (Yan et al, 2019). Therefore, we speculated that Dppa2/4 overexpression might accelerate the degradation of reprogramming factors in the oocyte cytoplasm, resulting in reprogramming failure. Furthermore, fertilized embryos are completely different from SCNT‐derived embryos, which are generated by directly injecting somatic nuclei into enucleated oocytes, whereas the oocyte cytoplasm is evolutionarily designed to reprogram spermatozoa. Therefore, although Dux and Dppa2/4 as ZGA inducers, their effects on the reprogramming process likely differ between sperm and somatic cells. As such, elucidation of relationships between Dux, Dppa2/4, and ZGA in SCNT warrants further investigation.
In summary, our study revealed that transient overexpression of Dux not only improved SCNT efficiency but also increased the efficiency of CiPSCs induction. Dux has been identified as a ZGA inducer in mice and humans, and it likely performed a similar function in all mammals (Iturbide & Torres‐Padilla, 2017; Whiddon et al, 2017). Therefore, these results suggest that transient overexpression of Dux might improve the somatic cell reprogramming efficiency of all mammals.
Materials and Methods
Animals and chemicals
Specific pathogen‐free‐grade B6D2F1 (C57BL/6 × DBA/2), CD1, and Kun‐Ming (KM) mice were purchased from Laboratory Animal Research Center (Inner Mongolia University) or Vital River Laboratories (Beijing, China). All animal experiments were approved by the Animal Care and Use Committee of Inner Mongolia University. All procedures were carried out in strict accordance with the recommendations made in the Guide for the Care and Use of Laboratory Animals of the National Veterinary and Quarantine Service. MERLV:: tdTomato transgenic mice (B6D2F1 background), which carry a transgenic MERLV promoter LTR‐driven tdTomato reporter (Yang et al, 2018), were mated with the same positive mice. dDux‐MEFs cells were isolated from E12.5 fetus. The MERLV:: tdTomato embryos used in the experiment were produced by MERVL::tdTomato sperm and MII oocytes from the littermates of transgenic mice. All chemicals used in this study were purchased from Sigma (USA), unless otherwise indicated.
SCNT and embryo culture
The SCNT processes followed previously published studies (Kishigami et al, 2006; Yang et al, 2018). Female B6D2F1 mice were used to provide somatic donor cells and oocytes, and CD‐1 or KM background mice were used as pseudopregnant surrogates. Due to the two‐step method is more harmful to SCNT embryos (too much micromanipulation), we adopted a one‐step micromanipulation technique to produce the SCNT embryos. Briefly, groups of ~ 50 MII oocytes were transferred to a chamber containing oil‐covered M2 supplemented with 5 μg/ml cytochalasin B (CB). The nuclei of donor cells were drawn in and out of the injection pipette until its plasma membrane was broken, and the donor cells were then sucked into the injection pipette. The MII oocyte spindle‐chromosome complex (SCC) was adjusted to 8 to 10 o'clock, and then, one donor cell was injected into the nearby plasma. The SCC was immediately aspirated into the injection pipette (~ 10 μm internal diameter) using the Piezo micromanipulator (PrimeTech, Japan) on a 37°C heating stage of an inverted microscope (Nikon, Japan). The reconstructed embryos were cultured in aMEM medium (Thermo, USA) containing 10% fetal calf serum (FCS; Hyclone, USA) for 1 h before activation treatment. The reconstructed embryos were activated in Ca2+‐free KSOM medium containing 10 mM strontium and 5 μg/ml CB for 6 h. Activated embryos were thoroughly washed and cultured in G1/G2 medium (1:1, vol/vol; Vitrolife, Sweden). Induction of Dux was performed by administration of doxycycline (2 μg/ml) in the G1/G2 medium.
Immunofluorescence staining
Embryos and CiPS cells were rinsed three times in phosphate‐buffered saline (PBS) with 0.3% BSA, fixed with 4% paraformaldehyde (PFA) overnight at 4°C, and then permeabilized with 0.2% (vol/vol) Triton X‐100 for 15 min at room temperature, followed by washing thoroughly in PBS containing 0.3% BSA. Fixed samples were blocked in 0.05% Tween‐20 in PBS containing 3% BSA (PBST) at 37°C for 1 h and then incubated with the primary antibodies overnight at 4°C. Samples were incubated with primary antibodies against Zscan4 (Millipore, AB4340, USA), MuERVL‐Gag (EpiGentek, A‐2801‐100, USA), Oct4 (Santa Cruz, sc‐8629, USA), and Ssea1 (Santa Cruz, sc‐21702, USA). After incubating, the samples were needed to wash several times in PBST and then incubated with appropriate secondary antibodies conjugated with Alexa Fluor 594 and Alexa Fluor 488 (Thermo, USA) for 1 h at 37°C. For imaging, the embryos were mounted in 10 μl anti‐fade solution with DAPI (Thermo, USA) and compressed with a cover‐slip. All samples were observed by a laser scanning microscope (Nikon, Japan).
In vitro mRNA synthesis and microinjection in oocytes
The Dux sequence was amplified from the pCW57.1‐mDux‐CA plasmid (Addgene 99284). The coding region of Dppa2 and Dppa4 was synthesized by BGI genomics institute (Beijing, China). The Dux, Dppa2, and Dppa4 were cloned into T7‐driven vectors, and mRNAs were synthesized in vitro using mMESSAGE mMACHINE T7 Ultra Kit (Thermo, USA). The final concentration of mRNA was diluted to 900 ng/ml before injection. As previously described (Yang et al, 2018), 8 pl of mRNA was microinjected into the cytoplasm of denuded oocytes. These oocytes were obtained by superovulating B6D2F1 female mice by intraperitoneal injection of 10 IU pregnant mare serum gonadotropin (PMSG; Sansheng, China) and 10 IU human chorionic gonadotropin (hCG; Sansheng, China), 48 h apart. The cumulus‐oocyte complexes (COCs) were collected from oviducts 14‐h post‐hCG, and the cumulus cells were dispersed by EmbryoMax FHM Mouse Embryo Media (Millipore, USA). Oocytes were injected using Piezo‐operated blunt‐end micropipette (3–5 μm internal diameter). After injection, oocytes were kept at room temperature for 30 min and then moved into the incubator. The siRNA information was presented in Appendix Table S5.
Generation of dox‐inducible Dux transgenic mice
The pCW57.1‐mDux‐CA vector was a gift from Stephen Tapscott (Addgene 99284). The pronuclear microinjection for the production of transgenic mice followed previously published studies (Ittner & Gotz, 2007). Briefly, the vector was injected into the well‐recognized pronuclei. Injected zygotes were transferred into pseudopregnant female mice (~ 30 zygotes per mouse) after 4‐h recovery culture in KSOM‐AA medium (Millipore, USA). For founder identification, tail tips were subjected to standard DNA extraction procedures. The amplified DNA fragments were subjected to TA cloning and sequencing. The founder mice were crossed to the littermates of founder mice for four generations to produce homozygous Dux mice.
Generation of Dux‐knockout mice via CRISPR/Cas9
The pronuclear microinjection was performed as previously described (Yang et al, 2014; Song et al, 2019). Briefly, B6D2F1 female mice were super‐ovulated through intraperitoneal (i.p.) injection of pregnant mare serum gonadotropin (10 IU; Sansheng, China) and human chorionic gonadotropin (hCG, 10 IU, China), 48 h apart. After the hCG i.p. injection, the female mice mated with male B6D2F1 mice in a 1:1 ratio in single cages overnight. The next day morning, the zygotes were collected from the oviducts. After purification, Cas9 mRNA (100 ng/μl) and sgRNAs (50 ng/μl each) were directly injected into zygotes. Following injection, zygotes were maintained at room temperature for 30 min and then moved into the incubator. The injected zygotes were transferred into pseudopregnant female mice (~ 30 zygotes per mouse) after a 2‐h recovery culture in KSOM‐AA medium (Millipore, USA). After 19–21 days, the mice pups were delivered naturally. For founder identification, the tail tips (~ 1 cm) were subjected to standard DNA extraction procedures. The amplified DNA fragments were subjected to TA cloning and sequencing. The founder (F0) mice were crossed with their littermates to produce F1 mice. The target sites of Dux gene were designed according to previously reported (De Iaco et al, 2017; Chen & Zhang, 2019), and sgRNA sequences were presented in Appendix Table S6.
Generation of CiPS cells
This section was adapted from Deng's laboratory (Zhao et al, 2018). Reagents setup: Small molecules: VPA, CHIR99021, 616452, Tranylcypromine, Forskolin, Ch55, EPZ004777, DZNep, Decitabine, SGC0946, and PD0325901. Stage I medium: FBS/KSR‐based medium supplemented with the small‐molecule cocktail VC6TF5E (100 μM VPA, 40 μM CHIR99021, 10 μM 616452, 5 μM Tranylcypromine, 10 μM Forskolin, 1 μM Ch55, and 5 μM EPZ004777). N2B27‐SII medium: N2B27‐based medium supplemented with 10 ng/ml LIF, 50 μg/ml vitamin C, 25 ng/ml bFGF, 2 mg/ml Albumax‐II, and the small‐molecule cocktail VC6TF5ZDS (VPA at 1 mM). Stage III medium: N2B27‐based medium with 3 μM CHIR99021, 1 μM PD0325901, 10 ng/ml LIF, and 50 μg/ml vitamin C. On day –1, MEFs were seeded at 50,000 cells per well of six‐well plate with MEF culture medium. The next day (day 0), change the medium into optimized stage I medium. From day 6–12, cells were cultured in optimized N2B27‐SII medium. During day 12–16, cells were cultured in stage III medium with 500 μM VPA. On day 16, VPA was removed and cells were cultured in stage III medium. After another 4–10 days, CiPS colonies emerged. Induction of Dux was performed by administration of doxycycline (2 μg/ml) in the culture medium.
Alkaline phosphatase staining
For alkaline phosphatase staining, CiPS cells were fixed with 4% PFA in PBS for 2 min, rinsed once with PBS and detection was performed using an Alkaline Phosphatase Assay Kit (VECTOR, USA) according to the manufacturer's protocol.
MTT cell viability assay
Cell viability was measured using the CyQUANT MTT Cell Viability Assay kit (Invitrogen, Carlsbad, CA, USA). The ESCs were cultured in 96‐well plates (2.0 × 103 cells/well). After 72 h culture, 10 μl 3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) stock solution (12 mM) was added to each well and incubated at 37°C for 4 h. After incubation, the supernatants were removed and 100 μl DMSO (Sigma, St. Louis, MO, USA) was added to each well. When the intracellular formazan crystals were solubilized at 37°C, the absorbance value was evaluated on a microplate reader (Thermo, Waltham, MA, USA) at a wavelength of 540 nm.
RT‐qPCR analysis
Total RNA was isolated using TRIzol reagent (Thermo, USA) and was immediately reverse‐transcribed using a Prime Script RT reagent kit (Takara, Japan). The reverse transcription PCR was amplified using Ex Taq (Takara, Japan). The RT‐qPCR was performed using an SYBR Premix Ex Taq (Takara, Japan), and signals were detected with Applied Biosystems 7500 real‐time PCR System (Thermo, USA). Relative mRNA expression was calculated to use the method. The single‐embryo RT‐qPCR was done as previously described (Yang et al, 2018; Yan et al, 2019), with some modifications. Briefly, embryonic total RNA was extracted using an RNeasy Micro Kit (Qiagen, Germany) and treated with DNase following the manufacturer's instructions. mRNAs were reversed by SuperScript III Reverse Transcriptase Kit (Thermo, USA). The expression levels of all embryos were normalized to the average expression levels of the ICSI group. The primer information was presented in Appendix Table S6.
Chimeric mice, chimeric blastocyst, and embryo transfer
For chimeric mice, CiPSCs were used 1 day before passaging, which showed an optimal undifferentiated morphology. The CiPSCs were microinjected into CD1/KM blastocysts using a Piezo (PrimeTech, Japan) microinjection pipette. After culturing for 3 h, the reconstructed blastocysts were transplanted into the uterus of pseudopregnant mice (~ 15 embryos per mouse). For chimeric blastocyst, four CiPSCs labeled by EGFP fluorescence were microinjected into 8‐cell stage B6D2F1 mouse embryos. The reconstructed embryos were cultured in ESCs medium for the first 5 h and then changed into the KSOM‐AA embryo culture medium (Millipore, USA) to obtain the chimeric blastocysts. For SCNT animals, the 2‐cell stage SCNT embryos were transferred to the oviducts of E0.5 pseudopregnant (~ 20 embryos per mouse). The cloned pups were nursed by lactating CD1/KM females. SSLP analysis was performed for D6Mit15 and D2Mit102. The primer information is presented in Appendix Table S6.
Teratoma formation
Teratoma formation analysis was carried out to evaluate the pluripotency of CiPS cells. Approximately 106 CiPS cells were injected subcutaneously into the hind limbs of 8‐week‐old nude mice. After 6 weeks, fully formed teratomas were dissected and fixed with PBS containing 4% PFA, then embedded in paraffin, sectioned, and stained with hematoxylin and eosin for histological analysis.
Bisulfite genomic sequencing
Genomic DNA was extracted by the tissue and blood DNA extraction kit (Qiagen, Germany) and treated with the Methylamp DNA modification kit (EpiGentek, USA). The bisulfite conversion was performed as previously published studies (Cao et al, 2018).
RNA sequencing
The single‐cell RNA‐seq method followed previously published studies (Tang et al, 2010; Liu et al, 2016). The reverse transcription was performed directly on the cytoplasmic lysate of individual embryos. The total cDNA library was then amplified by 18–20 cycles for library construction, which was performed following the manufacturer's instructions (Illumina, USA). Paired‐end sequencing was further performed at Annoroad (Beijing, China). Three biological replicates were analyzed for each treatment condition. After removing low‐quality reads and adapters, the raw reads were mapped to the mm9 genome using Tophat (v1.3.3) with default parameters (Trapnell et al, 2009). Expression levels for all RefSeq transcripts were quantified to fragments per kilobase of exon model per million mapped reads (FPKM) using Cufflinks (v1.2.0) (Trapnell et al, 2010). Gene Ontology (GO) analyses for differentially expressed genes were performed by using the R package GO.db and Database for Annotation, Visualization and Integrated Discovery (DAVID) (da Huang et al, 2009).
Statistics analysis
Statistical analyses were done using the univariate analysis of variance (ANOVA) followed by the Student t‐test with SPSS 21.0 statistical software (Armonk, USA). P < 0.05 was considered significant.
Author contributions
LY and GL conceived and designed the study. LY, XL, LS, AD, GS, CB, and ZW performed the experiments. LY, XL, LS, AD, and GS analyzed the data. GL supervised the project. LY and GL wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Movie EV1
Movie EV2
Review Process File
Acknowledgements
We thank Prof. Shaorong Gao (Tongji University), Prof. Zhiming Han (Chinese Academy of Sciences), and Prof. Qing Xia (Peking University) for their technical assistance. We also thank Prof. Yi Zhang (Harvard Medical School) for sharing the list of RRRs and Dux‐gRNA sequence. This study was supported by the Genetically Modified Organisms Breeding Major Projects (2016ZX08007‐002), the opening project of State Key Laboratory of R2BGL (to LY), the Inner Mongolia University Chief Scientist Program (to GL), the Inner Mongolia Autonomous Region Basic Research Project (to GL).
EMBO Reports (2020) 21: e50054
Data availability
Sequencing data have been deposited in the NCBI sequence read archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession IDs: SCNT‐1 (SRR10201281), SCNT‐2 (SRR10201280), SCNT‐3 (SRR10201279); D‐SCNT‐1 (SRR10201278), D‐SCNT‐2 (SRR10201277), D‐SCNT‐3 (SRR10201285); ICSI‐1 (SRR10201284), ICSI‐2 (SRR10201283), ICSI‐3 (SRR10201282); D‐CiPS‐1 (SRR11453944), D‐CiPS‐2 (SRR11453943), D‐CiPS‐3 (SRR11453942); CiPS‐1 (SRR11453941), CiPS‐2 (SRR11453940), CiPS‐3 (SRR11453939).
References
- Ancelin K, Syx L, Borensztein M, Ranisavljevic N, Vassilev I, Briseno‐Roa L, Liu T, Metzger E, Servant N, Barillot E et al (2016) Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. Elife 5: e08851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha‐Mohammadi S, Tassin A, Coppee F et al (2008) An isogenetic myoblast expression screen identifies DUX4‐mediated FSHD‐associated molecular pathologies. EMBO J 27: 2766–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Daughters RS, Xu Z, Slack JM, Kyba M (2009) Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD‐related repeats. PLoS ONE 4: e7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, da Silva MT, Sunny ST, Ener ET, Toso EA, Yuan C, Cui Z, Walters MA, Jadhav A, Kyba M (2019) A novel P300 inhibitor reverses DUX4‐mediated global histone H3 hyperacetylation, target gene expression, and cell death. Sci Adv 5: eaaw7781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Yu S, Li D, Ye J, Yang X, Li C, Wang X, Mai Y, Qin Y, Wu J et al (2018) Chromatin accessibility dynamics during chemical induction of pluripotency. Cell Stem Cell 22: 529–542 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang Y (2019) Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet 51: 947–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, Kyba M (2016) DUX4 recruits p300/CBP through its C‐terminus and induces global H3K27 acetylation changes. Nucleic Acids Res 44: 5161–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Risso D, Chen S, Kim TA, Tan MH, Li JB, Wu Y, Chen C, Xuan Z et al (2017) Deficiency of microRNA miR‐34a expands cell fate potential in pluripotent stem cells. Science 355: eaag1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D (2017) DUX‐family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Coudray A, Duc J, Trono D (2019) DPPA2 and DPPA4 are necessary to establish a 2C‐like state in mouse embryonic stem cells. EMBO Rep 20: e47382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramskold D, Reinius B, Sandberg R (2014) Single‐cell RNA‐seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196 [DOI] [PubMed] [Google Scholar]
- Eckersley‐Maslin MA, Alda‐Catalinas C, Reik W (2018) Dynamics of the epigenetic landscape during the maternal‐to‐zygotic transition. Nat Rev Mol Cell Biol 19: 436–450 [DOI] [PubMed] [Google Scholar]
- Eckersley‐Maslin M, Alda‐Catalinas C, Blotenburg M, Kreibich E, Krueger C, Reik W (2019) Dppa2 and Dppa4 directly regulate the Dux‐driven zygotic transcriptional program. Genes Dev 33: 194–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl JO, Giesige CR, Domire JS, Wallace LM, Fowler AM, Guckes SM, Garwick‐Coppens SE, Labhart P, Harper SQ (2016) Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum Mol Genet 25: 4577–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Tian CL, Ye X, Sheng X, Wang H, Liu Y, Liu L (2018) Dynamics of telomere rejuvenation during chemical induction to pluripotent stem cells. Stem Cell Reports 11: 70–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Wu X, Djekidel MN, Zhang Y (2019) Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat Cell Biol 21: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Zheng J, Xing F, Fang H, Sun F, Yan A, Gong X, Ding H, Tang F, Sheng HZ (2007) Nuclear reprogramming: the strategy used in normal development is also used in somatic cell nuclear transfer and parthenogenesis. Cell Res 17: 135–150 [DOI] [PubMed] [Google Scholar]
- Gao R, Wang C, Gao Y, Xiu W, Chen J, Kou X, Zhao Y, Liao Y, Bai D, Qiao Z et al (2018) Inhibition of aberrant DNA re‐methylation improves post‐implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell 23: 426–435 [DOI] [PubMed] [Google Scholar]
- Guo M, Zhang Y, Zhou J, Bi Y, Xu J, Xu C, Kou X, Zhao Y, Li Y, Tu Z et al (2019) Precise temporal regulation of Dux is important for embryo development. Cell Res 29: 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL et al (2017) Conserved roles of mouse DUX and human DUX4 in activating cleavage‐stage genes and MERVL/HERVL retrotransposons. Nat Genet 49: 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R (2003) Nuclear transplantation, embryonic stem cells, and the potential for cell therapy. N Engl J Med 349: 275–286 [DOI] [PubMed] [Google Scholar]
- da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez‐Gasca R, Mizutani E, Boskovic A, Ziegler‐Birling C, Rodriguez‐Terrones D, Wakayama T, Vaquerizas JM, Torres‐Padilla ME (2015) Early embryonic‐like cells are induced by downregulating replication‐dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671 [DOI] [PubMed] [Google Scholar]
- Ittner LM, Gotz J (2007) Pronuclear injection for the production of transgenic mice. Nat Protoc 2: 1206–1215 [DOI] [PubMed] [Google Scholar]
- Iturbide A, Torres‐Padilla ME (2017) Starting embryonic transcription for the first time. Nat Genet 49: 820–821 [DOI] [PubMed] [Google Scholar]
- Kishigami S, Wakayama S, Thuan NV, Ohta H, Mizutani E, Hikichi T, Bui HT, Balbach S, Ogura A, Boiani M et al (2006) Production of cloned mice by somatic cell nuclear transfer. Nat Protoc 1: 125–138 [DOI] [PubMed] [Google Scholar]
- Liu W, Liu X, Wang C, Gao Y, Gao R, Kou X, Zhao Y, Li J, Wu Y, Xiu W et al (2016) Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single‐cell sequencing. Cell Discov 2: 16010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M et al (2018) Cloning of macaque monkeys by somatic cell nuclear transfer. Cell 172: 881–887 [DOI] [PubMed] [Google Scholar]
- Lu F, Zhang Y (2015) Cell totipotency: molecular features, induction, and maintenance. Natl Sci Rev 2: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y (2014) Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159: 884–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Zhang Y (2018) Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell 23: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML (1995) Regulation of gene expression at the beginning of mammalian development. J Biol Chem 270: 22077–22080 [DOI] [PubMed] [Google Scholar]
- Nothias JY, Miranda M, DePamphilis ML (1996) Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO J 15: 5715–5725 [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut‐Karslioglu A, Biechele S, Huang B, Shen X, Ramalho‐Santos M (2018) A LINE1‐Nucleolin partnership regulates early development and ESC identity. Cell 174: 391–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WM 3rd, Hochedlinger K, Kyba M, Daley GQ, Jaenisch R (2002) Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell 109: 17–27 [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Osorio N, Urrego R, Cibelli JB, Eilertsen K, Memili E (2012) Reprogramming mammalian somatic cells. Theriogenology 78: 1869–1886 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard‐Liesching H, Verp S, Marquis J et al (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Schultz RM (1993) Regulation of zygotic gene activation in the mouse. BioEssays 15: 531–538 [DOI] [PubMed] [Google Scholar]
- Song L, Yang L, Wang J, Liu X, Bai L, Di A, Li G (2019) Generation of Fad2 and Fad3 transgenic mice that produce n‐6 and n‐3 polyunsaturated fatty acids. Open Biol 9: 190140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA (2010) RNA‐seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 5: 516–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA‐seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA‐seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao C, Hou Z, Yang Y, Bi Y, Wang H, Zhang Y, Gao S (2018) Unique molecular events during reprogramming of human somatic cells to induced pluripotent stem cells (iPSCs) at naive state. Elife 7: e29518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson JA, Simon AK, Myrick DA, Wolf G, Driscoll S, Pfaff SL, Macfarlan TS, Katz DJ (2016) Maternally provided LSD1/KDM1A enables the maternal‐to‐zygotic transition and prevents defects that manifest postnatally. Elife 5: e08848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ (2017) Conservation and innovation in the DUX4‐family gene network. Nat Genet 49: 935–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML (1991) Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol 147: 403–414 [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810–813 [DOI] [PubMed] [Google Scholar]
- Xu X, Smorag L, Nakamura T, Kimura T, Dressel R, Fitzner A, Tan X, Linke M, Zechner U, Engel W et al (2015) Dppa3 expression is critical for generation of fully reprogrammed iPS cells and maintenance of Dlk1‐Dio3 imprinting. Nat Commun 6: 6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YL, Zhang C, Hao J, Wang XL, Ming J, Mi L, Na J, Hu X, Wang Y (2019) DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS Biol 17: e3000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Jaenisch R (2014) Generating genetically modified mice using CRISPR/Cas‐mediated genome engineering. Nat Protoc 9: 1956–1968 [DOI] [PubMed] [Google Scholar]
- Yang L, Song L, Liu X, Bai L, Li G (2018) KDM6A and KDM6B play contrasting roles in nuclear transfer embryos revealed by MERVL reporter system. EMBO Rep 19: e46240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu X, Song L, Su G, Di A, Bai C, Wei Z, Li G (2019) Inhibiting repressive epigenetic modification promotes telomere rejuvenation in somatic cell reprogramming. FASEB J 33: 13982–13997 [DOI] [PubMed] [Google Scholar]
- Yang F, Huang X, Zang R, Chen J, Fidalgo M, Sanchez‐Priego C, Yang J, Caichen A, Ma F, Macfarlan T et al (2020) DUX‐miR‐344‐ZMYM2‐mediated activation of MERVL LTRs induces a totipotent 2C‐like state. Cell Stem Cell 26: 234–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Fu Y, Zhu J, Liu Y, Zhang Q, Yi Z, Chen S, Jiao Z, Xu X, Xu J et al (2018) Single‐cell RNA‐seq reveals dynamic early embryonic‐like programs during chemical reprogramming. Cell Stem Cell 23: 31–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Movie EV1
Movie EV2
Review Process File
Data Availability Statement
Sequencing data have been deposited in the NCBI sequence read archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession IDs: SCNT‐1 (SRR10201281), SCNT‐2 (SRR10201280), SCNT‐3 (SRR10201279); D‐SCNT‐1 (SRR10201278), D‐SCNT‐2 (SRR10201277), D‐SCNT‐3 (SRR10201285); ICSI‐1 (SRR10201284), ICSI‐2 (SRR10201283), ICSI‐3 (SRR10201282); D‐CiPS‐1 (SRR11453944), D‐CiPS‐2 (SRR11453943), D‐CiPS‐3 (SRR11453942); CiPS‐1 (SRR11453941), CiPS‐2 (SRR11453940), CiPS‐3 (SRR11453939).


