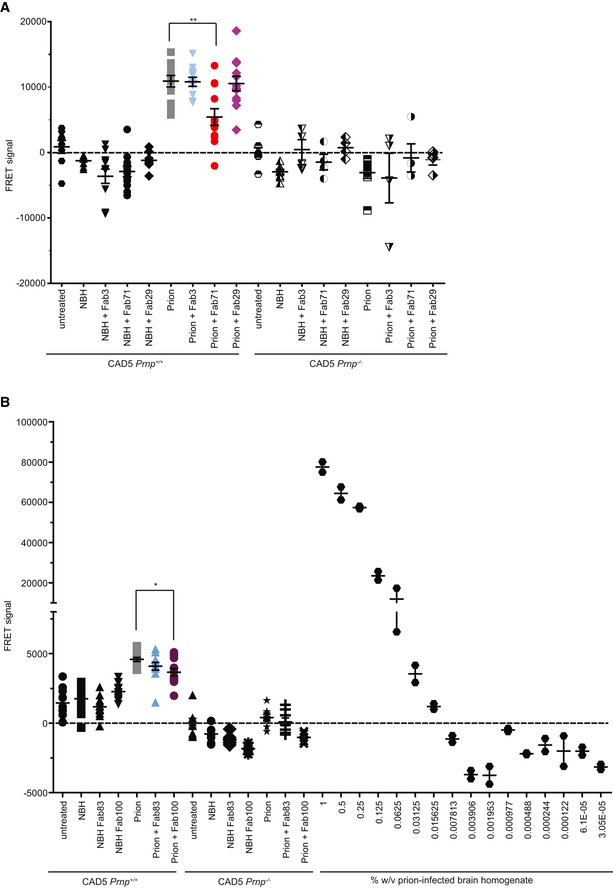
Figure EV5. TR‐FRET for the detection of wtPrPS c in prion‐infected CAD5 Prnp +/+ after treatment with a panel of Fabs.
-
ATR‐FRET for the detection of PrPSc in prion‐infected CAD5 Prnp +/+ cells after treatment with selected anti‐PrP Fabs. Treatment of the cells with Fab71 (OR51‐91), but not with Fab3 (CC123‐50) or Fab29 (GD), significantly reduced PrPSc levels compared to untreated, prion‐infected cells.
-
BTreatment of prion‐infected CAD5 Prnp +/+ cells with Fab100 (OR51‐91 binder), but not with Fab83 (CC123‐50 binder), significantly reduced wtPrPSc levels compared to untreated prion‐infected CAD5 Prnp +/+ cells. The dilution range (3 × 10−5% to 1% w/v) of prion‐infected BH is shown on the right site to indicate the linear range of the assay. Negative controls: NBH and CAD5 Prnp −/− cells. Here and in (A), the FRET signal of untreated CAD5 Prnp −/− cells was set to zero. The FRET anti‐PrP antibody pair POM19‐Eu and POM1‐APC was used for detection.
Data information: For prion‐infected CAD5 Prnp +/+ cells, the assay was performed in biological triplicates represented as four technical replicates for Fab71, Fab3, Fab100, and Fab83. For CAD5 Prnp +/+ cells that were either untreated, exposed to NBH, or treated with Fab29, the assay was performed in biological duplicates represented as four technical replicates. For CAD5 Prnp −/− cells, one biological replicate was assayed as four technical replicates. n = 4–12. Each dilution of prion‐infected BH is represented in technical duplicates. Data presented in dot plots represent the mean ± sem. One‐way ANOVA and Bonferroni's multiple comparisons were used for statistical analysis; *P < 0.05; **P < 0.01.
