Figure EV1. USP44 is up‐regulated in Tregs and largely tracked with the progressive elevation of Treg‐associated transcript levels.
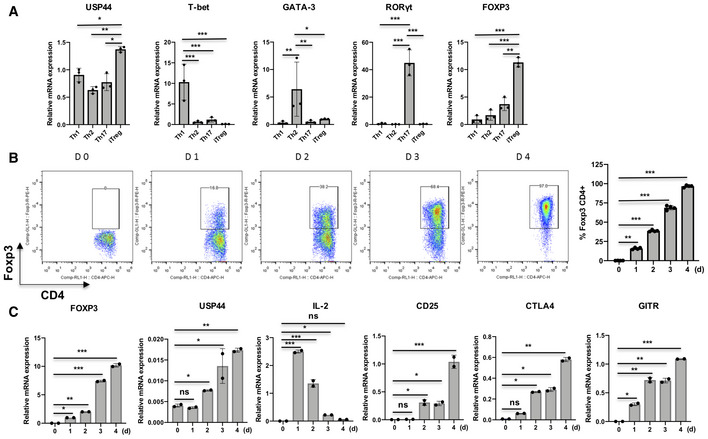
- Naïve CD4+ T cells were isolated from the peripheral blood of healthy donors (n = 2–3/experiment) and differentiated into the indicated T helper lineages by activation (anti‐CD3/anti‐CD28 antibodies; 1 and 4 μg/ml, respectively) in the presence of specific skewing reagents (described in the Materials and Methods section). Cells were harvested after 4 days, and mRNA was isolated for qRT–PCR analysis of USP44 message and that of key lineage‐defining transcription factors.
- iTregs were generated by in vitro skewing as above, and cells were harvested at different time points. Flow cytometry confirmed the progressive up‐regulation of FOXP3 under iTreg skewing conditions, and bar graph was displayed.
- qRT–PCR analysis revealed the levels of FOXP3, USP44, IL‐2, CD25, CTLA‐4, and GITR encoding transcripts in iTregs.
