-
A, B
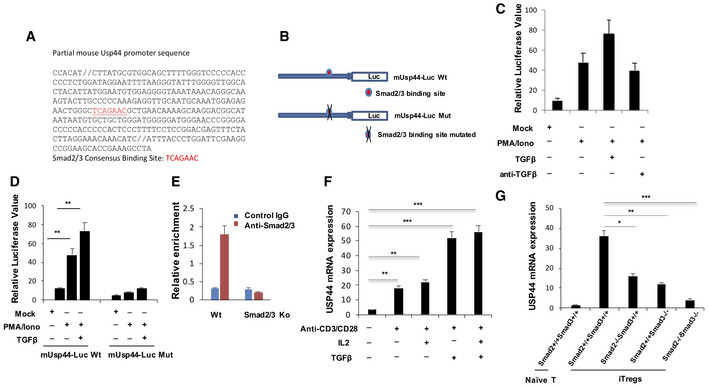
The USP44 promoter sequence is represented with its SMAD binding site indicated. The effect of SMAD2/3 binding site loss was found by making the mutations depicted in (B).
-
C
Jurkat T cells were transfected with an USP44 promoter‐driven firefly luciferase reporter by electroporation. Cells were stimulated with PMA and ionomycin in the presence or absence of TGF‐β (2 ng/ml) or anti‐TGF‐β (10 μg/ml) for 8 h before being harvested for measurement of luciferase activity, which was normalized to Renilla luciferase activity.
-
D
Jurkat T cells were transfected with an USP44 promoter‐driven firefly luciferase reporter by electroporation. Cells were stimulated with PMA and ionomycin in the presence or absence of TGF‐β (2 ng/ml) for 8 h before being harvested for measurement of luciferase activity, which was normalized to Renilla luciferase activity.
-
E
ChIP analysis of SMAD occupancy of the Usp44 promoter. Naïve T cells were isolated from C57BL/6 mice and polarized to iTreg before harvest and lysis. SMAD2 and SMAD3 factors were immunoprecipitated, and PCR was used to detect binding to the Usp44 promoter sequence.
-
F
FACS isolated naïve T cells were exposed to the indicated stimuli during ex vivo culture and were harvested at different time points for mRNA isolation and measurement of USP44 expression by qRT–PCR analysis.
-
G
Naïve CD4+ T cells were isolated from wild‐type mice (Smad2+/+Smad3+/+) as well as mice lacking SMAD2 (Smad2−/−Smad3+/+), SMAD3 (Smad2+/+Smad3−/−), or both SMAD2 and SMAD3 (Smad2/3−/−), all on a C57BL/6 background (n = 3/group/experiment). Cells were activated under in vitro Treg‐inducing conditions (anti‐CD3/CD28, 1 and 4 μg/ml, respectively) in the presence of IL‐2 (100 U/ml) and TGF‐β (5 ng/ml) for 4 days. Then, RNA was extracted, and cDNA was prepared in order to assess USP44 expression levels in these cells by qRT–PCR.
Data information: Panels (C–G) depict mean results from three independent experiments (biological replicates) ± SEM. *
‐test.