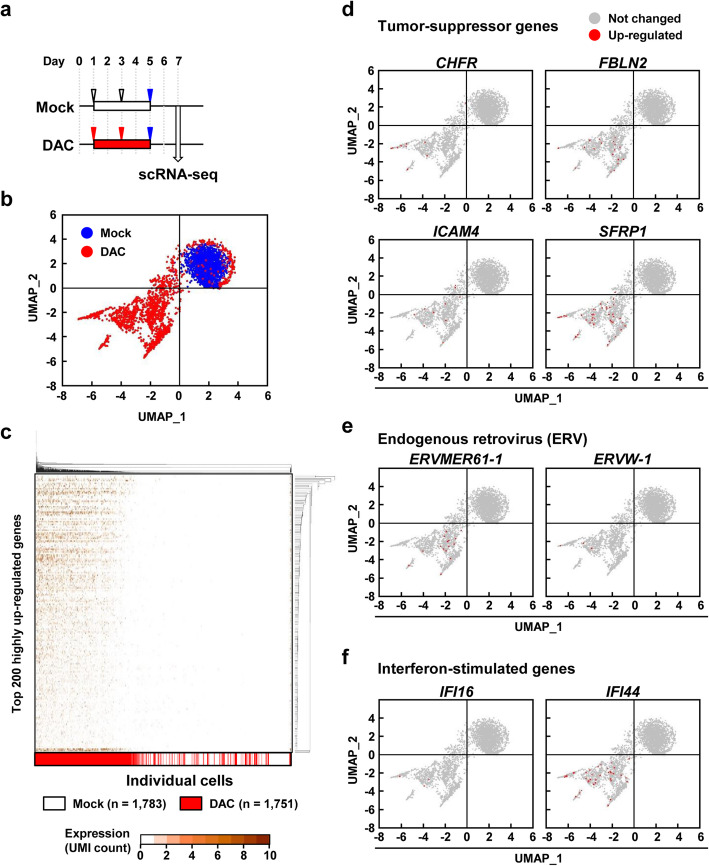
Fig. 1.
scRNA-seq of DAC-treated HCT116 cells. a Experimental protocol of DAC treatment. HCT116 cells were seeded on day 0 and treated with 0.5 μM of DAC on days 1 and 3. The cells were placed in fresh medium without DAC on day 5, and harvested on day 7. b UMAP using 14,099 genes that can be induced by DAC treatment. DAC-treated HCT116 cells had diverse expression profiles at the single-cell level. Blue dots, mock-treated cells. Red dots, DAC-treated cells. c Unsupervised hierarchical clustering analysis using the top 200 highly upregulated genes. Genes with higher expression levels were different, depending upon DAC-treated clones. White, mock-treated cells. Red, DAC-treated cells. d, e, and f Upregulation of specific tumor-suppressor genes, ERVs, and interferon-stimulated genes. Tumor-suppressor gene methylation silenced in colorectal cancers (CHFR, FBLN2, ICAM4, and SFRP1), ERVs (ERVMER61-1 and ERVW-1), and interferon-stimulated genes (IFI16 and IFI44) were upregulated in random fractions of cells