Abstract
BACKGROUND/OBJECTIVES
Genetic factors play an important role in Alzheimer’s disease (AD) and cognitive aging. However, it is unclear whether risk loci identified in European ancestry (EA) populations have similar effects in other groups, such as South Asians.
DESIGN
We investigated the allelic distribution and cognitive associations of 56 known AD risk single-nucleotide polymorphisms (SNPs) identified from three EA genome-wide association studies (EA-GWASs) in a South Asian population. Single SNP and genetic risk score (GRS) associations with measures of episodic memory were assessed.
SETTING
The Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India (LASI-DAD).
PARTICIPANTS
A total of 906 LASI-DAD participants from diverse states in India.
MEASUREMENTS
Participants were genotyped using the Illumina Global Screening Array and imputed with 1000G Phase 3v5. Cognitive measures included total learning and delayed word recall.
RESULTS
Although only a few SNPs were significantly associated with memory scores (P < .05), effect estimates from the EA-GWAS and the LASI-DAD showed moderate correlation (0.35–0.88) in the expected direction. GRSs were also associated with memory scores, although percentage variation explained was small (0.1%–0.6%).
CONCLUSIONS
Discrepancies in allele frequencies and cognitive association results suggest that genetic factors found predominantly through EA-GWASs may play a limited role in South Asians. However, the extent of differences in the genetic architecture of AD and cognition in EA and South Asians remains uncertain. There is also a critical need to perform a more comprehensive assessment of the mutational spectrum of South Asia to identify novel genetic variants associated with AD and cognition in this population.
Keywords: Alzheimer’s disease, cognition, genetics, South Asian, Indian
INTRODUCTION
Alzheimer’s disease (AD) is a progressive form of dementia with pronounced impairment in memory. Recently, genetic factors have been linked to AD, and the estimated heritability of AD is as high as 80%.1–3 The strongest genetic risk factor is the ε4 allele of the apolipoprotein E (APOE) gene, with an odds ratio (OR) of 14.9 in individuals who carry two ε4 alleles versus noncarriers.4,5 However, the ε2 allele of APOE is protective from AD.6,7
Genome-wide association studies (GWASs) have identified other genetic risk factors for AD. The first large-scale GWAS of AD, conducted by Lambert and colleagues using 74,046 AD cases and controls, identified 19 AD risk loci in addition to APOE.8 Recently, Kunkle et al expanded the discovery cohort (N = 94,437) and identified 25 AD risk loci in total, 7 of which were new.9 A third GWAS, conducted by Jansen et al,10 added UK Biobank data and included both clinically diagnosed AD and AD-by-proxy (through parental diagnosis) cases (N = 455,258). They identified 29 risk loci, including 12 that were novel.10 Taken together, these AD GWASs identified 66 unique AD-associated single-nucleotide polymorphisms (SNPs) from 38 loci in addition to APOE (Supplementary Table S1).
Importantly, all three AD GWASs were conducted among European ancestry (EA) partcipants.8–10 Currently, there are no large-scale AD GWASs in Indian or South Asian populations. This is problematic because populations may have different linkage disequilibrium (LD) patterns due to their unique evolutionary history, potentially resulting in genetic heterogeneity across ancestries.11 Most South Asian groups descended from a mixture of two genetically divergent populations: ancestral North Indians related to Central Asians, Middle Easterners, Caucasians, and Europeans; and ancestral South Indians not related to any groups outside of the subcontinent.12,13 Following the mixture, a major demographic shift toward endogamy led to strong founder events more extreme than in Ashkenazi Jews or Finns.13,14 As a result, LD structure and allele frequencies differ substantially not only between South Asian and European populations, but also across neighboring groups in India.15 Further, genetic predictors of dementia may differ between EA and South Asian populations due to differences in environmental factors (including diet, toxicants, or socioeconomic status), which may interact with or overshadow genetic risk factors. Therefore, is not clear whether genetic loci previously identified in EA-GWASs will perform similarly in South Asians.
To assess the transferability of genetic risk variants discovered through EA-based AD GWASs to South Asians, we evaluated the effect of these variants with cognitive function in participants of the Longitudinal Aging Study in India–Diagnostic Assessment of Dementia (LASI-DAD). We tested 66 unique AD-associated SNPs from three GWASs to present an inclusive set of SNPs, representing both highly replicated, larger-effect AD risk variants 8 and newly discovered, smaller-effect AD risk variants from studies with larger sample sizes and/or broader case definitions.9,10 We also evaluated the effect of APOE ε4 and ε2 alleles. Both single SNP association and genetic risk score (GRS) associations were evaluated.
METHODS
Study Population
The LASI is a nationally representative sample of greater than 70,000 adults from India aged 45 years or older. LASI-DAD, an add-on study, includes the Harmonized Cognitive Assessment Protocol (HCAP), informational interviews, and blood draws for approximately 3,000 LASI respondents aged 60 years or older. To guarantee a sufficient number of respondents with dementia and mild cognitive impairment, a stratified random sample design was implemented.16
Genotyping and Imputation
The Illumina Infinium Global Screening Array-24 (GSA) BeadChip, version 2.0 (Illumina) was used to genotype 960 LASI-DAD participants. Genotypes were imputed to the 1000G Project worldwide reference panel (phase 3, version 5). Principal component (PC) analysis17 was used to exclude outliers and select a set of 932 unrelated individuals for analysis. The top 10 PCs were included in all analyses to adjust for population stratification. Supplemental Methods provides additional details.
Cognitive Measures and Covariates
A battery of cognitive tests was administered, including a common set of cognitive tests from the HCAP16 to enable international comparisons and additional cognitive tests suitable for illiterate and innumerate populations. Cognitive measures analyzed were total learning score (0–30 words) and delayed word recall score (0–10 words). Cognitive tests and surveys were translated into 10 local languages. Education level was categorized as less than lower secondary education, upper secondary education/vocational training, or tertiary education. A total of 906 individuals had genotype data, covariate data (sex, age, and education), and at least one cognitive measurement.
Association Between SNPs and Cognitive Function
We compared the allelic distribution of the AD risk SNPs between LASI-DAD and the EA-GWAS samples by calculating the correlation coefficient of the risk allele frequency across all SNPs. We assessed whether allele frequency of each SNP differed between LASI-DAD and EA-GWAS samples using a one-sample proportion test.
Linear regression was used to assess whether each SNP was associated with total learning and/or delayed recall scores in LASI-DAD, using two models. Model 1 adjusted for age, sex, and the top 10 genetic PCs, and model 2 additionally adjusted for education. We hypothesized that AD risk alleles would be associated with lower cognitive function. We assessed results at a nominal significance level (P ≤ .05) as well as a Bonferroni-corrected significance level, accounting for the number of SNPs analyzed. We assessed the correlation of the risk allele effect sizes on cognitive function in LASI-DAD with the effect sizes on AD reported from the three AD GWASs. We further tested SNP-by-age interactions to evaluate whether effect sizes vary by age.
Association Between GRSs and Cognitive Function
Three GRSs were constructed using all the identified AD risk SNPs from each AD GWAS separately. Note that there is overlap in some of the SNPs that comprise these three scores. We excluded variants in the APOE region from the three GRSs and treated APOE as an independent signal. Each GRS was calculated as , with βi being the effect size associated with the risk allele for SNP i, and xij being the dosage of the risk allele for SNP i in individual j. The effect size of each SNP was calculated as the ln(OR) reported in the corresponding GWAS article. We assessed whether each GRS was associated with total learning or delayed recall using the regression models above. We then combined the three GRSs into a single multivariable model to assess the total variance in cognitive function explained before and after adding APOE ε2 and ε4.
Sensitivity Analysis of the APOE Region
Because APOE is known to be the most important risk locus for AD, we also tested the association between all SNPs in APOE (plus 2 kb upstream to capture the promoter region) and memory scores. For SNPs associated with any memory score, we tested for interaction with ε4 and ε2.
RESULTS
Descriptive Statistics
The LASI-DAD sample was 44% male, and study participants were 69 years old (standard deviation (SD) = 7.2 years) on average (Table 1). Most participants attained lower than secondary level education (70.0%), and over half were from a rural area. Participants had a mean of 12.0 and 3.4 words (SD = 5.2 and 2.4 words) for total learning and delayed recall scores, respectively. The correlation between memory scores was r = 0.73 (P < .0001).
Table 1.
Characteristics of the LASI-DAD Sample (N = 906)
| Characteristic | Mean or No. | SD or % |
|---|---|---|
| Age, y | 69.16 | 7.22 |
| Male sex | 400 | 44.2 |
| Total learning score (words)a | 12.01 | 5.15 |
| Delayed recall score (words)b | 3.35 | 2.43 |
| Education | ||
| Less than lower secondary | 634 | 70.0 |
| Upper secondary/vocational training | 233 | 25.7 |
| Tertiary | 39 | 4.3 |
| Caste | ||
| 1. Scheduled caste | 170 | 18.8 |
| 2. Scheduled tribe | 17 | 1.9 |
| 3. Other backward class | 450 | 49.7 |
| 4. None of them | 269 | 29.7 |
| Urban/rural | ||
| Urban community | 400 | 44.2 |
| Rural village | 506 | 55.8 |
Abbreviation: LASI-DAD, Longitudinal Aging Study in India–Diagnostic Assessment of Dementia.
Total learning score, N = 899.
Delayed recall score, N = 893.
Genotype and Imputation Quality in LASI-DAD
Among those genotyped, the median call rate was excellent (99.95%), and the estimated error rate was low (1.5 × 10−6). The mean EmpRsq (correlation between the true genotypes and imputed dosages calculated by masking the given SNP) was 0.86 for common variants (minor allele frequency [MAF] >0.05), indicating relatively high quality, but was lower (0.66) for rare variants (MAF ≤0.05, comprising 84.7% of the measured variants).
Allelic Distribution of the AD Risk Loci
Among the 68 unique SNPs, 27 were directly genotyped on the GSA, and another 29 were successfully imputed with high quality (r2 > 0.8). One SNP was not available because it was not included in the 1000G reference panel, and another 12 SNPs were excluded due to poor imputation quality in LASI-DAD. As a result, 56 unique SNPs were investigated (Supplementary Tables S1 and S2).
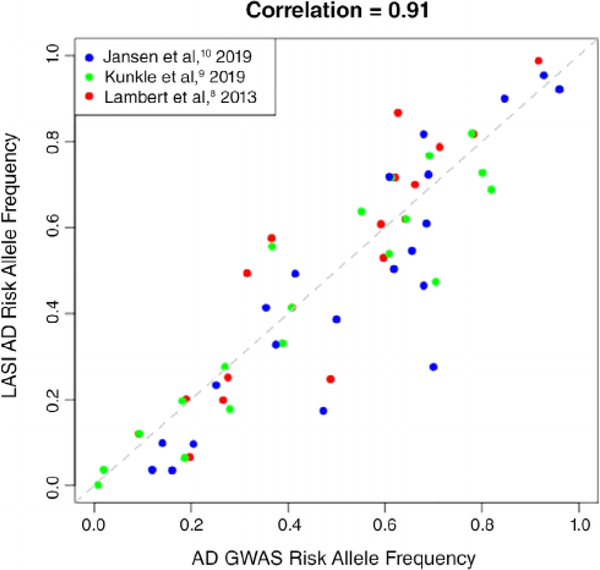
Figure 1 compares the risk allele frequencies of all 56 unique SNPs in LASI-DAD versus the corresponding AD GWAS. Although the risk allele frequencies are correlated as expected (r = 0.91; P < .0001), 47 SNPs had a significantly different allele frequency between LASI-DAD and the AD GWAS samples (P ≤ .05; Supplementary Table S3). We note that this frequency comparison will be slightly biased toward the null because GWAS studies are best powered to detect more common variants. Thus, the SNPs identified in GWAS would have less variability in allele frequencies compared with an unselected set of SNPs. The variance ratio of the risk allele frequencies in the GWAS samples and LASI-DAD is 0.81, slightly lower than 1. Thus, the influence of the bias, if it exists, is relatively small.
Figure 1.
Scatterplot of the risk allele frequencies from Alzheimer’s disease (AD) genome-wide association study (GWAS) and Longitudinal Aging Study in India–Diagnostic Assessment of Dementia (LASI-DAD) sample. The risk allele frequencies of the 60 AD-associated single-nucleotide polymorphisms (SNPs) (56 unique SNPs and 3 SNPs that appeared in multiple articles) in LASI-DAD (y axis) are compared with the corresponding risk allele frequencies reported in the 3 AD GWASs (x axis). As shown, the risk allele frequencies of SNPs in LASI-DAD correlated well with those from the European ancestry AD GWAS samples (correlation = 0.91). The risk allele frequencies of APOE v2 and ε4 variants are taken from Jansen et al.10
Association Between Single SNPs and Cognitive Measures
Associations between each AD risk SNP and each cognitive measure were assessed using two models. Model 1 adjusted for age, sex, and the top 10 genetic PCs, and model 2 additionally adjusted for education. Q-Q plots indicate that in both models, P values from the 56 SNPs were generally smaller than expected by chance alone (Supplementary Figure S1). This suggests some evidence of association between the AD risk SNPs and cognitive measures.
All nominally significant association results (P ≤ .05) are presented in Table 2. Of the 56 SNPs assessed, 10 were nominally associated with total learning and/or delayed recall score (Table 2). Among the 10 SNPs, 3 (rs2830500 (ADAMTS1), rs10948363 (CD2AP6), and rs11218343 (SORL1)) were associated with both memory scores in model 1. Two SNPs (rs1859788 (ZCWPW1) and rs7185636 (IQCK)) were associated with total learning score only, and one SNP (rs9473117 (CD2AP)) was associated with delayed recall score only in model 1. Significance was attenuated for rs2830500 (ADAMTS1) and rs1859788 (ZCWPW1) for total learning score after controlling for education (model 2). In contrast, four SNPs (rs4147929 and rs2752246 in ABCA7 and rs6733839 and rs4663105 in BIN1) show a stronger association with total learning and/or delayed recall in model 2 than model 1. However, no associations remained significant after Bonferroni correction (P ≤ 8.9 × 10−4). Among the 10 SNPs, most demonstrated an association in the expected direction, where AD risk alleles were associated with lower memory score. Neither the APOE ε2 nor ε4 allele was associated (P ≤ .05) with memory scores, although the effect directions were as expected.
Table 2.
SNPs with at Least One Nominally Significant Association in LASI-DAD (P ≤ .05), and APOE ε2 and ε4 Alleles
| SNP | Gene | GWAS study | Risk allele | Risk AF | Chr | Position | Total Learning Score |
Delayed Recall Score |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 |
Model 2 |
Model 1 |
Model 2 |
|||||||||||
| β | P value | β | P value | β | P value | β | P value | |||||||
| rs2830500 | ADAMTS1 | Kunkle et al9 | C | 0.77 | 21 | 28,156,856 | −.66 | .019 | −.5 | .059 | −.44 | .001 | −.38 | .003 |
| rs10948363 | CD2AP | Lambert et al8 | G | 0.20 | 6 | 47,487,762 | .62 | .025 | .52 | .041 | .4 | .002 | .35 | .004 |
| rs9473117 | CD2AP | Kunkle et al9 | C | 0.18 | 6 | 47,431,284 | .57 | .064 | .48 | .091 | .4 | .006 | .36 | .009 |
| rs4147929 | ABCA7 | Lambert et al8 | A | 0.20 | 19 | 1,063,443 | .55 | .062 | .67 | .015 | .24 | .095 | .29 | .03 |
| rs3752246 | ABCA7 | Kunkle et al9 | G | 0.20 | 19 | 1,056,492 | .51 | .098 | .59 | .037 | .2 | .169 | .24 | .078 |
| rs1859788 | ZCWPW1 | Jansen et al10 | G | 0.72 | 7 | 99,971,834 | −.59 | .018 | −.41 | .072 | −.1 | .418 | −.03 | .817 |
| rs6733839 | BIN1 | Lambert et al8 and Kunkle et al9 | T | 0.41 | 2 | 127,892,810 | −.43 | .059 | −.44 | .034 | −.18 | .096 | −.19 | .065 |
| rs4663105 | BIN1 | Jansen et al10 | C | 0.49 | 2 | 127,891,427 | −.39 | .078 | −.43 | .036 | −.18 | .092 | −.20 | .050 |
| rs7185636 | IQCK | Kunkle et al9 | T | 0.69 | 16 | 19,808,163 | −.59 | .017 | −.53 | .021 | −.10 | .379 | −.08 | .445 |
| rs11218343 | SORL1 | Lambert et al,8 Kunkle et al,9 and Jansen et al10 | T | 0.92 | 11 | 121,435,587 | −.83 | .046 | −.89 | .02 | −.56 | .005 | −.58 | .002 |
| rs429358 | APOE | ε4 | C | 0.10 | 19 | 45,411,941 | −.30 | .438 | −.20 | .572 | −.15 | .383 | −.12 | .487 |
| rs7412 | APOE | ε2 | C | 0.95 | 19 | 45,412,079 | −.49 | .36 | −.19 | .702 | −.17 | .516 | −.04 | .877 |
Note: P ≤ .05 is indicated by bold text. Effects sizes (β values) are calculated with respect to the Alzheimer’s disease risk allele.
Abbreviations: AF, allele frequency; Chr, chromosome; GW AS, genome-wide association study; LASI-DAD, Longitudinal Aging Study in India–Diagnostic Assessment of Dementia; SNP, single-nucleotide polymorphism.
Although only a small number of the 56 SNPs were significant, most had the expected effect directions (62.5% for total learning and 57.1% for delayed recall in model 1; Supplementary Table S4). AD risk alleles were generally associated with decreased cognition in LASI-DAD (r between effect estimates ranges from −0.35 to −0.88; Supplementary Table S5, Supplementary Figure S2). SNPs with larger GWAS effect sizes tended to have the expected direction of effect in our study; however, those with smaller effect sizes did not always have the expected effect direction. Thus, the relatively strong correlation may not necessarily reflect consistent effects of AD risk variants on cognition.
Among 56 tested SNPs, 6 interacted with age at P ≤ .05 in one or multiple models (Supplementary Table S6), but none was significant after Bonferroni correction, suggesting that the SNP effects do not vary substantially by age.
Association Between GRSs and Cognitive Measures
The distribution of the three GRSs (constructed from three AD GWASs) are presented in Supplementary Figure S3. As expected, the GRSs were positively correlated with each other (Table 3). However, none of the correlations was strong, suggesting that they capture similar but distinct genetic risk profiles in LASI-DAD.
Table 3.
Correlations of GRSs Calculated from Alzheimer’s Disease GWASs
| Correlation | GRS (Lambert et al8) |
GRS (Kunkle et al9) |
GRS (Jansen et al10) |
|---|---|---|---|
| GRS (Lambert et al8) | 1.000 | ||
| GRS (Kunkleetal9) | 0.798 | 1.000 | |
| GRS (Jansen et al10) | 0.567 | 0.592 | 1.000 |
Note: All GRSs are standardized to a N(0,1) distribution.
Abbreviations: GRS, genetic risk score; GWAS, genome-wide association study.
In regression model 1, the Lambert et al8 GRS and the Kunkle et al9 GRS were negatively associated with total learning score (Table 4), and the Kunkle et al9 GRS was negatively associated with delayed recall score. In model 2, the Kunkle et al9 GRS was associated with total learning score, but all other GRSs were attenuated. The proportion of variance in total learning score explained by each GRS ranged from 0.2% to 0.6% in model 1 and from 0.1% to 0.4% in model 2. When combined together, the three GRSs explained 0.7% and 0.4% of variance in model 1 and model 2, respectively. When further combined with APOE ε4 and ε2, they explained 0.8% in model 1 and 0.4% in model 2. Compared with total learning score, the proportion of variance in delayed recall score explained by each GRS showed a similar pattern but was smaller.
Table 4.
GRS Associations with Cognitive Function
| Total learning score |
Delayed recall score |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 |
Model 2 |
Model 1 |
Model 2 |
|||||||||
| Article | β | P value | R2, % | β | P value | R2, % | β | P value | R2, % | β | P value | R2, % |
| GRS (Lambert et al8) | −.313 | .050 | 0.4 | −.252 | .089 | 0.2 | −.094 | .219 | 0.1 | −.066 | .361 | 0.1 |
| GRS (Kunkleetal9) | −.420 | .008 | 0.6 | −.317 | .032 | 0.4 | −.179 | .018 | 0.5 | −.136 | .059 | 0.3 |
| GRS (Jansen et al10) | −.215 | .178 | 0.2 | −.145 | .330 | 0.1 | −.068 | .376 | 0.1 | −.039 | .588 | 0.02 |
| GRS (Lambert et al8) + GRS (Kunkle et al9) + GRS (Jansen et al10) | 0.7 | 0.4 | 0.7 | 0.4 | ||||||||
| GRS (Lambert et al8) + GRS (Kunkle et al9) + GRS (Jansen et al10) + rs7412(ε2) + rs429358 (ε4) | 0.8 | 0.4 | 0.8 | 0.4 | ||||||||
Note: P ≤ .05 is indicated by bold text. GRS effects sizes (β values) are calculated with respect to an increasing number of Alzheimer’s disease risk alleles. Model 1: cognitive measure ~ GRS + sex + age + principal component (PC) 1 − PC10. Model 2: cognitive measure ~ GRS + sex + age + education + PC1 − PC10. R2 represents the variance in the cognitive measure explained by the individual GRS, three GRSs combined, or GRSs combined with APOE variants. Abbreviation: GRS, genetic risk score.
Sensitivity Analysis of the APOE Region
There were 10 SNPs within APOE (plus the 2-kb promoter) region in LASI-DAD. Among the 10 SNPs, all 3 SNPs in the promoter region (rs7256173, rs7259620, and rs405509) and 2 SNPs within the gene (rs440446 and rs769450) were nominally associated with total learning score in models 1 and/or 2 at P ≤ .05 (Supplementary Table S7). Two SNPs (rs7259620 and rs440446) remained significant after Bonferroni correction (P ≤ .005).
We further tested the interaction of these SNPs with both rs429358 (ε4) and rs7412 (ε2). We observed a nominally significant interaction (P ≤ .05) between rs7256173 and rs429358 (ε4) for both total learning and delayed recall scores in models 1 and 2, and a significant interaction between rs440446 and rs7412 (ε2) on delayed recall score in model 2 (Supplementary Table S8). The interaction between rs7256173 and rs429358 (ε4) on total learning score remained significant after Bonferroni correction (P ≤ .01). Suggestive evidence of interactions (P < .1) were observed for rs405509 by rs429358 (ε4) on total learning score in model 2, rs440446 by rs7412 (ε2) on both memory scores in models 1 and 2, and rs405509 by rs7412 (ε2) for both memory scores in model 2.
DISCUSSION
We investigated the association between AD-associated SNPs identified from EA-GWASs and cognitive function in a South Asian cohort. Many of the AD risk variants had different allele frequencies in LASI-DAD compared with EA-GWAS samples. This could be due to differences in study design. Neither LASI-DAD nor the EA-GWASs are fully population-representative. EA-GWAS samples are mostly case-control studies, and LASI-DAD is a substudy of the population-representative LASI study, with oversampling those at high risk for cognitive impairment. On the other hand, there could be true differences in allele frequency between the two populations. This would not be surprising due to strong founder events in Indian/South Asian populations.18 Differences in LD patterns between European and Indian/South Asian ancestries may also cause different SNPs at the same genes/loci to be more strongly associated with AD in each population. Further, because Indians/South Asians derive more than half of their ancestry from a founding population that no longer exists in an unmixed form and is significantly divergent from other extant populations, it is unlikely that all of the variants identified in the EA populations are directly transferrable to Indian/South Asian populations.11,14
Genetic predictors of dementia may also differ between populations when there are major differences in environmental risk factors. Dementia and cognitive decline may result from AD-related neurodegeneration, but also from other common disease processes, such as cardiovascular disease. The prevalence of hypertension has increased rapidly in India due to longer life expectancy and westernization in lifestyle.19 Previous studies indicate a larger burden of vascular dementia over AD in Asian populations, whereas AD-related dementia is more prevalent in EA.20 Also, LASI-DAD participants differ in other ways from EA-GWAS participants, including diet (e.g., enriched with curcumin spice), natural environment (e.g., greater exposure to various pollutants), and social environment (e.g., lower socioeconomic status and education levels). These environmental factors may be associated with dementia/cognition alone and/or through interaction with genetic risk factors.21–25 Thus, the underlying genetic risk factors may differ in these populations. It is therefore critical to validate the effects of AD risk variants in Indians/South Asians.
Although India is the second largest country in the world, it is rarely represented in genomic studies.12 The 1000 Genomes Project only contains a small proportion (13.6%) of Indians/South Asians, and they are not well represented in any other commonly used large reference panel, including the Trans-Omics for Precision Medicine and the Haplotype Reference Consortium.26 As a result, some of the SNPs had low imputation quality in LASI-DAD, and thus could not be evaluated in this study. Imputation quality was especially low for rare variants, as has been previously demonstrated.27 Therefore, large-scale, population-based Indian/South Asian sequencing projects are needed to attain a representative reference panel that would allow for higher-quality imputation for future Indian/South Asian-based studies and facilitate efforts to identify new variants linked to AD and other diseases.
This study suggests that EA-GWAS AD risk variants confer a small amount of genetic risk for decreased cognitive function in Indians/South Asians. Therefore, previously identified AD risk variants may not be the most prominent or the only risk variants in Indians/South Asians. At a nominal P value, risk variants in ADAMTS1, ZCWPW1, IQCK, BIN1, and SORL1 were associated with decreased cognitive function in LASI-DAD. Among them, BIN1, SORL1, and ZCWPW1 were the early identified risk loci in Lambert et al.8 BIN1 is one of the most strongly associated loci for AD after APOE. The gene is expressed primarily in the central nervous system and is believed to activate a caspase-independent apoptotic process.28 ZCWPW1 is a histone modification reader and potentially involved in phosphoinositide 3-kinase signaling pathways in neurons.29 The association between both loci and AD has been reported in East Asians, although the implicated SNPs were not the same.29,30 Interestingly, the variant we examined at the ZCWPW1 locus (rs1859788) is a missense mutation and has been recently suggested to likely be a causal allele for AD.31 SORL1 encodes a type I transmembrane protein that helps facilitate lipid reception through endocytosis and sorting of fats. The T allele of rs1121834 in SORL1 was associated with increased risk of late-onset AD in East Asians.32,33 The suggestive evidence of associations observed in LASI-DAD suggest that these loci are likely to have similar function in Indians/South Asians and EA.
ADAMTS1 and IQCK were novel loci that were identified by the recent AD GWASs (ADAMTS1 in both Kunkle et al and Jansen et al10; IQCK in Kunkle et al9). ADAMTS1 encodes a zinc-binding enzyme excreted in several adult tissues that is required for normal ovulation and renal function,34 and has been linked to breast cancer.35 IQCK encodes a protein with an IQ motif that allows for binding of EF-hand proteins and has been linked to obsessive compulsive disorder.36 Additional studies are needed to understand the roles of these genes in AD and cognition.
Interestingly, two SNPs from CD2AP and two SNPs from ABCA7 were nominally associated with increases in total learning and delayed recall scores. CD2AP encodes a scaffolding protein that supports the integrity of intercellular junctions. ABCA7 encodes a multispan transmembrane protein that is highly expressed in the brain, facilitating transport of phospholipids and cholesterol across cell membranes.37 Associations between SNPs in these two genes and AD in East Asian cohorts have shown mixed results.38,39 Similar to our study, a study in approximately 850 Koreans found that several AD risk variants in Europeans were actually protective for AD at a nominal significance level, including the A allele of rs4147932, which also shows the unexpected direction of effect in our study as well.40 Thus, CD2AP and ABCA7 likely play a role in AD and/or cognition in Indians/South Asians, although the involved risk variants may differ from EA. We also noted that none of the results from these genes remained significant after multiple testing correction in our study, and thus should be interpreted cautiously.
The two APOE major isoforms were not associated with memory scores in LASI-DAD, although the effect estimates were in the expected direction. APOE is the strongest genetic risk locus for AD in multiple populations, including Asians.41 Two recent meta-analyses show that APOE ε4 is associated with AD in Indians, with similar effect sizes as in EA (OR ranging from 4.14 to 5.90).42,43 Interestingly, one study found that APOE ε3 instead of ε2 showed suggestive evidence of protection against AD in Indians.43 The lack of association between APOE major isoforms and cognition in this study may be due to our examination of memory scores rather than diagnosed AD, which are not necessarily good predictors of AD risk.44,45 Prior studies found that the association between APOE and memory performance/cognition was much weaker than with AD.46,47 In addition, the association may be age dependent, with stronger associations at older ages.48–50 Further, ε4 is more strongly associated with story-based than word-based memory tasks.48 Consistent with our study, multiple studies in Asians failed to find an association between APOE variants and cognition.51,52 Although poor episodic memory/cognition is a key symptom of AD, it may also be a consequence of other health conditions, such as Huntington’s disease, Parkinson’s disease, hypertension, and others.53–56 Thus, lack of association for APOE major isoforms, and possibly for other AD risk variants, may be due to the outcomes investigated in this study.
In addition to APOE major isoforms, many studies suggest that the variants in the promotor region and intron 1, in particular rs405509 and rs440446, were associated with cognitive function/AD independently or through interaction with the major APOE isoforms.57–60 Consistent with this literature, the same SNPs along with three other variants (two in the promoter region) were associated with total learning score in this study. Furthermore, we observed a significant interaction between a promoter SNP and ε4 on both total learning score and delayed recall score as well as suggestive evidence of interaction that includes both rs405509 and rs440446. This evidence suggests that APOE is probably an important risk locus for dementia in Indians/South Asians as well, but that multiple variants and mechanisms might be involved. Nonetheless, given that we investigated memory rather than AD, there is still tremendous uncertainty regarding the effect of these genetic risk factors on AD in South Asians. Future studies that focus on clinically diagnosed AD in this population will be the essential next step to compare the SNP effect sizes across populations and better understand the genetic architecture of AD in South Asians.
Although most AD risk variants were not strongly associated with cognitive function, GRSs composed of these variants did show association with cognitive function in LASI-DAD. The GRS constructed from Kunkle et al9 performed best among all three GRSs, possibly because this study is the best-powered GWAS with clinically diagnosed AD cases. Although Jansen et al10 have a muc h larger sample size, the case definition for this GWAS included both clinically diagnosed AD as well as AD by proxy (parental diagnosis). The additional cases identified through proxy may have introduced bias or noise to the analysis, reducing predictive ability of the GRS from this GWAS. Another contributing factor may be that approximately one-third of the risk variants from Jansen et al10 were from novel loci with potentially small and hard-to-replicate effects. Thus, the GRS from Jansen et al10 may have lower signal/noise ratio compared with other GRSs. The combined GRSs plus APOE ε4 and ε2 accounted for less than 1% of the variance in LASI-DAD memory scores. This explains drastically less than the 16% AD variance explained by AD risk variants in EA (APOE ε4 and ε2 explained 13%, whereas other genes explained 3%).53 Given the low variance explained in LASI-DAD, we caution against applying EA-based genetic knowledge to Indians/South Asians in a clinical setting.
In summary, this was the first study to comprehensively survey all AD risk variants identified in three large-scale AD GWASs and examine their association with cognitive function in an Indian/South Asian population. This study demonstrated some evidence of association between AD risk variants and lower cognition, although the effect sizes were small. A GRS of the known AD risk variants explained less than a percentage of the total variance in cognitive function, suggesting that many AD risk loci in Indians/South Asians may remain to be identified. Therefore, future large-scale AD GWASs and whole genome sequence analysis in Indian/South Asian samples are warranted to better characterize and understand the genetic cause of AD in this population.
Supplementary Material
Supplementary Figure S1: Q-Q plots for the association between cognitive measures and 56 known AD risk SNPs in LASI-DAD.
Supplementary Figure S2: Scatterplots of estimated effect sizes for cognitive measures in LASI-DAD and Alzheimer’s disease (AD) from published GWAS.
Supplementary Figure S3: Histograms of the genetic risk scores (GRSs) in LASI-DAD.
Supplementary Table S1: Alzheimer’s Disease Risk SNPs from Three Large, Replicated GWASs
Supplementary Table S2: Summary of Known Alzheimer’s Disease Risk SNPs in LASI-DAD
Supplementary Table S3: Comparison of Observed Risk Allele Frequency in LASI-DAD Versus Reported Risk Allele Frequency from Published Alzheimer’s Disease GWASs
Supplementary Table S4: SNP Associations with Total Learning Score and Delayed Recall Score
Supplementary Table S5: Correlations Between Effect Sizes from LASI-DAD and Alzheimer’s Disease GWASs
Supplementary Table S6: SNP-by-Age Interactions on Total Learning Score and Delayed Recall Score
Supplementary Table S7: Associations Between SNPs in the APOE Gene/Promoter Region and Total Learning or Delayed Recall Score
Supplementary Table S8: Interactions Between SNPs and APOE e2/e4 on Total Learning Score or Delayed Recall Score
ACKNOWLEGEMENTS
Financial Disclosure: This project is funded by the National Institute on Aging (R01 AG051125 and RF1 AG055273). The National Institute on Aging had no role in preparing the data or the manuscript.
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose.
Sponsor’s Role: The study sponsors had no role in the design and conduct of the study; in the collection, analysis, and interpretation of the data; or in the preparation, review, or approval of the manuscript.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Breteler MMB. Vascular risk factors for Alzheimer’s disease: an epidemiologic perspective. Neurobiol Aging. 2000;21:153–160. [DOI] [PubMed] [Google Scholar]
- 2.Bird TD. Genetic factors in Alzheimer’s disease. N Engl J Med. 2005;352: 862–864. [DOI] [PubMed] [Google Scholar]
- 3.Gatz M, Pedersen NL, Berg S, et al. Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52: M117–M125. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S. Molecular mechanism of apolipoprotein E binding to lipoprotein particles. Biochemistry. 2009;48:3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C-C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 2013;9:106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. [DOI] [PubMed] [Google Scholar]
- 7.Conejero-Goldberg C, Gomar JJ, Bobes-Bascaran T, et al. APOE2 enhances neuroprotection against Alzheimer’s disease through multiple molecular mechanisms. Mol Psychiatry. 2014;19:1243–1250. [DOI] [PubMed] [Google Scholar]
- 8.Lambert J-C, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin AR, Gignoux CR, Walters RK, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sengupta D, Choudhury A, Basu A, Ramsay M. Population stratification and underrepresentation of Indian subcontinent genetic diversity in the 1000 genomes project dataset. Genome Biol Evol. 2016;8:3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moorjani P, Thangaraj K, Patterson N, et al. Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93:422–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bittles AH. Population stratification and genetic association studies in South Asia . J Mol Genet Med. 2005;1:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Banerjee J, Khobragade PY, Angrisani M, Dey AB. LASI-DAD study: a protocol for a prospective cohort study of late-life cognition and dementia in India . BMJ Open. 2019;9:e030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakatsuka N, Moorjani P, Rai N, et al. The promise of discovering population-specific disease-associated genes in South Asia. Nat Genet. 2017;49:1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair T Challenges of hypertension and dementia in the Indian subcontinent: a review. J Hum Hypertens. 2019;33:568–574. [DOI] [PubMed] [Google Scholar]
- 20.Catindig JA, Venketasubramanian N, Ikram MK, Chen C. Epidemiology of dementia in Asia: insights on prevalence, trends and novel risk factors. J Neurol Sci. 2012;321:11–16. [DOI] [PubMed] [Google Scholar]
- 21.Peters R, Ee N, Peters J, Booth A, Mudway I, Anstey KJ. Air pollution and dementia: a systematic review. J Alzheimers Dis. 2019;70:S145–S163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulick ER, Elkind MSV, Boehme AK, et al. Long-term exposure to ambient air pollution, APOE-ε4 status, and cognitive decline in a cohort of older adults in northern Manhattan. Environ Int 2020;136:105440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seifan A, Schelke M, Obeng-Aduasare Y, Isaacson R. Early life epidemiology of Alzheimer’s disease–a critical review. Neuroepidemiology. 2015;45:237–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moorman SM, Carr K, Greenfield EA. Childhood socioeconomic status and genetic risk for poorer cognition in later life. Soc Sci Med. 2018;212:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy PH, Manczak M, Yin X, et al. Protective effects of Indian spice curcumin against amyloid-β in Alzheimer’s disease. J Alzheimers Dis. 2018;61:843–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S, Abecasis GR, Browning BL. Genotype imputation from large reference panels. Annu Rev Genomics Hum Genet. 2018;19:73–96. [DOI] [PubMed] [Google Scholar]
- 28.Lee JH, Cheng R, Barral S, et al. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. 2011;68:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Tan MS, Wang HF, et al. ZCWPW1 is associated with late-onset Alzheimer’s disease in Han Chinese: a replication study and meta-analyses. Oncotarget. 2016;7:20305–20311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan MS, Yu JT, Jiang T, Zhu XC, Guan HS, Tan L. Genetic variation in BIN1 gene and Alzheimer’ s disease risk in Han Chinese individuals. Neurobiol Aging. 2014;35:1781.e1–1788.e1. [DOI] [PubMed] [Google Scholar]
- 31.Rathore N, Ramani SR, Pantua H, et al. Paired immunoglobulin-like type 2 receptor alpha G78R variant alters ligand binding and confers protection to Alzheimer’s disease. PLoS Genet 2018;14:e1007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashita A, Koike A, Jun G, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C-C, Wang H-F, Tan M-S, et al. SORL1 is associated with the risk of late-onset Alzheimer’s disease: a replication study and meta-analyses. Mol Neurobiol. 2017;54:1725–1732. [DOI] [PubMed] [Google Scholar]
- 34.Lee NV, Sato M, Annis DS, et al. ADAMTS1 mediates the release of anti-angiogenic polypeptides from TSP1 and 2. EMBO J. 2006;25:5270–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malvia S, Bagadi SAR, Pradhan D, et al. Study of gene expression profiles of breast cancers in Indian women. Sci Rep. 2019;9:10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen BJ, Mills JD, Takenaka K, Bliim N, Halliday GM, Janitz M. Characterization of circular RNAs landscape in multiple system atrophy brain. J Neurochem. 2016;139:485–496. [DOI] [PubMed] [Google Scholar]
- 37.Fehér Á, Juhász A, Pákáski M, Janka Z, Kálmán J. Association study of the ABCA7 rs3752246 polymorphism in Alzheimer’s disease. Psychiatry Res. 2019;279:376–377. [DOI] [PubMed] [Google Scholar]
- 38.Tan L, Yu JT, Zhang W, et al. Association of GWAS-linked loci with late-onset Alzheimer’s disease in a northern Han Chinese population. Alzheimers Dement 2013;9:546–553. [DOI] [PubMed] [Google Scholar]
- 39.Xiao Q, Liu Z-J, Tao S, et al. Risk prediction for sporadic Alzheimer’s disease using genetic risk score in the Han Chinese population. Oncotarget. 2015;6:36955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung SJ, Lee JH, Kim SY, et al. Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer Dis Assoc Disord 2013;27:250–257. [DOI] [PubMed] [Google Scholar]
- 41.Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 42.Shankarappa BM, Kota LN, Purushottam M, et al. Effect of CLU and PIC-ALM polymorphisms on AD risk: a study from south India. Asian J Psychiatr 2017;27:7–11. [DOI] [PubMed] [Google Scholar]
- 43.Agarwal R, Tripathi CB. Association of apolipoprotein E genetic variation in Alzheimer’s disease in Indian population: a meta-analysis. Am J Alzheimers Dis Other Demen. 2014;29:575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanoiu A, Adam S, Van der Linden M, et al. Memory evaluation with a new cued recall test in patients with mild cognitive impairment and Alzheimer’s disease. J Neurol. 2005;252:47–55. [DOI] [PubMed] [Google Scholar]
- 45.Rabin LA, Paré N, Saykin AJ, et al. Differential memory test sensitivity for diagnosing amnestic mild cognitive impairment and predicting conversion to Alzheimer’s disease. Neuropsychol Dev Cogn B Aging Neuropsychol Cogn. 2009;16:357–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deary IJ, Whiteman MC, Pattie A, et al. Cognitive change and the APOE epsilon 4 allele. Nature. 2002;418:932. [DOI] [PubMed] [Google Scholar]
- 47.Small BJ, Rosnick CB, Fratiglioni L, Bäckman L. Apolipoprotein E and cognitive performance: a meta-analysis. Psychol Aging 2004;19:592–600. [DOI] [PubMed] [Google Scholar]
- 48.Debette S, Ibrahim Verbaas CA, Bressler J, et al. Genome-wide studies of verbal declarative memory in nondemented older people: the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Biol Psychiatry. 2015;77:749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu F, Pardo LM, Schuur M, et al. The apolipoprotein E gene and its age-specific effects on cognitive function. Neurobiol Aging. 2010;31:1831–1833. [DOI] [PubMed] [Google Scholar]
- 50.Mondadori CR, de Quervain DJ, Buchmann A, et al. Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers. Cereb Cortex. 2007;17:1934–1947. [DOI] [PubMed] [Google Scholar]
- 51.Yuan L, Liu J, Dong L, et al. Effects of APOE rs429358, rs7412 and GSTM1/GSTT1 polymorphism on plasma and erythrocyte antioxidant parameters and cognition in old Chinese adults. Nutrients. 2015;7:8261–8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shahar S, Lee LK, Rajab N, et al. Association between vitamin A, vitamin E and apolipoprotein E status with mild cognitive impairment among elderly people in low-cost residential areas. Nutr Neurosci. 2013;16:6–12. [DOI] [PubMed] [Google Scholar]
- 53.Ridge PG, Hoyt KB, Boehme K, et al. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol Aging. 2016;41:200–e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pillon B, Deweer B, Agid Y, Dubois B. Explicit memory in Alzheimer’s, Huntington’s, and Parkinson’s diseases. Arch Neurol. 1993;50:374–379. [DOI] [PubMed] [Google Scholar]
- 55.Aronow WS. Hypertension and cognitive impairment. Ann Transl Med. 2017;5:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker KA, Power MC, Gottesman RF. Defining the relationship between hypertension, cognitive decline, and dementia: a review. Curr Hypertens Rep. 2017;19:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rantalainen V, Lahti J, Henriksson M, et al. APOE and aging-related cognitive change in a longitudinal cohort of men. Neurobiol Aging. 2016;44:151–158. [DOI] [PubMed] [Google Scholar]
- 58.Ma C, Zhang Y, Li X, et al. Is there a significant interaction effect between apolipoprotein E rs405509 T/T and ε4 genotypes on cognitive impairment and gray matter volume? Eur J Neurol. 2016;23:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Limon-Sztencel A, Lipska-Ziętkiewicz BS, Chmara M, et al. The algorithm for Alzheimer risk assessment based on APOE promoter polymorphisms. Alzheimers Res Ther. 2016;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi KY, Lee JJ, Gunasekaran TI, et al. APOE promoter polymorphism-219T/G is an effect modifier of the influence of APOE ε4 on Alzheimer’s disease risk in a multiracial sample. J Clin Med. 2019;8:749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Q-Q plots for the association between cognitive measures and 56 known AD risk SNPs in LASI-DAD.
Supplementary Figure S2: Scatterplots of estimated effect sizes for cognitive measures in LASI-DAD and Alzheimer’s disease (AD) from published GWAS.
Supplementary Figure S3: Histograms of the genetic risk scores (GRSs) in LASI-DAD.
Supplementary Table S1: Alzheimer’s Disease Risk SNPs from Three Large, Replicated GWASs
Supplementary Table S2: Summary of Known Alzheimer’s Disease Risk SNPs in LASI-DAD
Supplementary Table S3: Comparison of Observed Risk Allele Frequency in LASI-DAD Versus Reported Risk Allele Frequency from Published Alzheimer’s Disease GWASs
Supplementary Table S4: SNP Associations with Total Learning Score and Delayed Recall Score
Supplementary Table S5: Correlations Between Effect Sizes from LASI-DAD and Alzheimer’s Disease GWASs
Supplementary Table S6: SNP-by-Age Interactions on Total Learning Score and Delayed Recall Score
Supplementary Table S7: Associations Between SNPs in the APOE Gene/Promoter Region and Total Learning or Delayed Recall Score
Supplementary Table S8: Interactions Between SNPs and APOE e2/e4 on Total Learning Score or Delayed Recall Score