Figure 3: TRAP-isolated RNA can be used for downstream gene expression analysis.
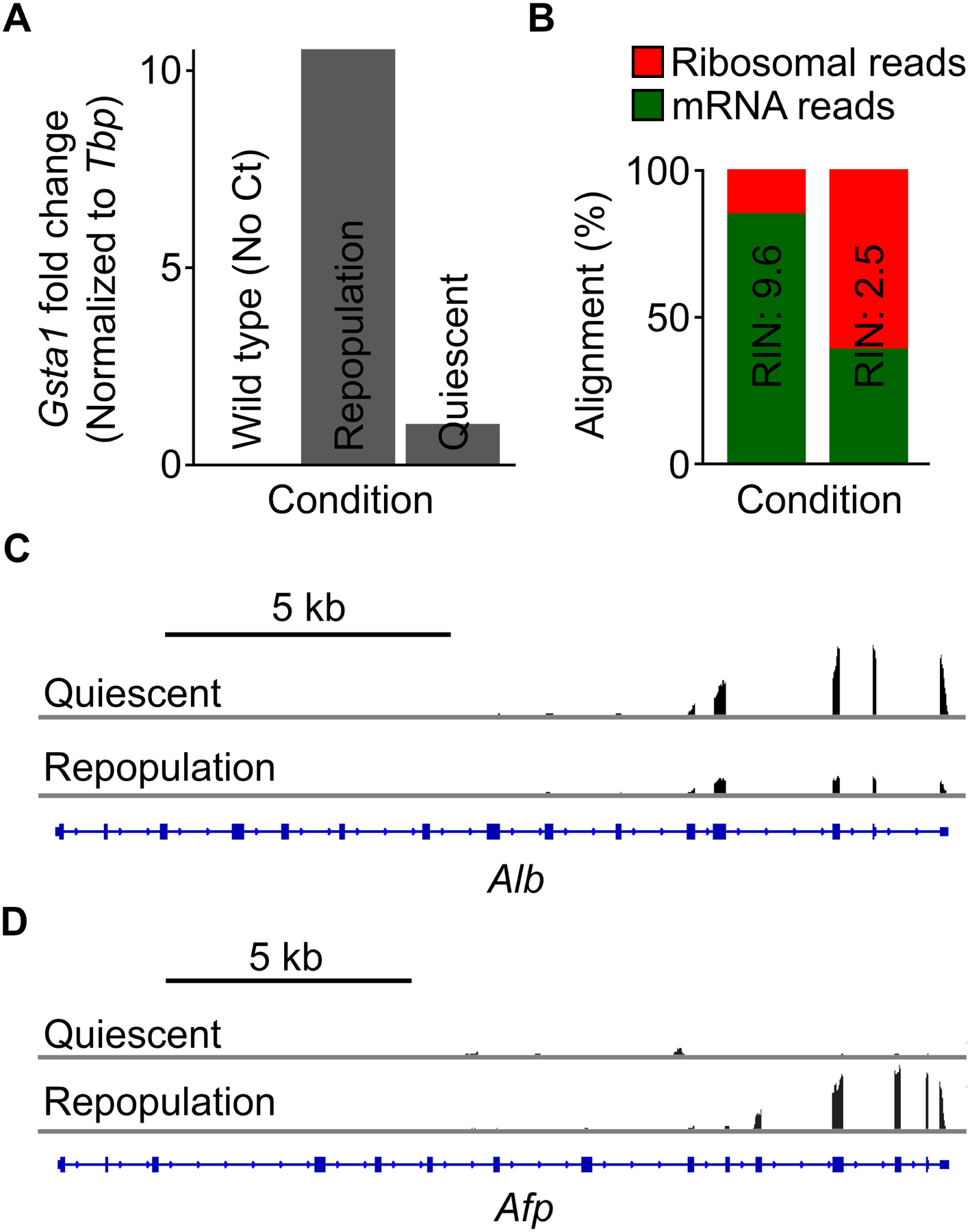
(A) Representative reverse transcription and quantitative PCR results of Gsta1 in quiescent and repopulating hepatocytes. No Ct value was detected with RNA isolated from wild type animals. (B) Alignment analysis of isolated RNA after high-throughput sequencing, demonstrating the importance of determining RNA integrity after isolation. High-quality RNA results in a higher percentage of mRNA reads (green), while low-quality RNA leads to a much higher percentage of ribosome reads (red), as most mRNA is degraded. RIN, RNA integrity number. (C,D) Integrative Genomics Viewer (IGV) tracks of RNA-seq reads of mRNA affinity-purified from quiescent and repopulating hepatocytes at the (C) Alb and (D) Afp loci. Note that the 3’ read bias is typical of a poly(A) selection pipeline.
