Figure 1.
Comparison of AI-ETD versus ETD and EThcD for Mapping ADP-Ribosylation
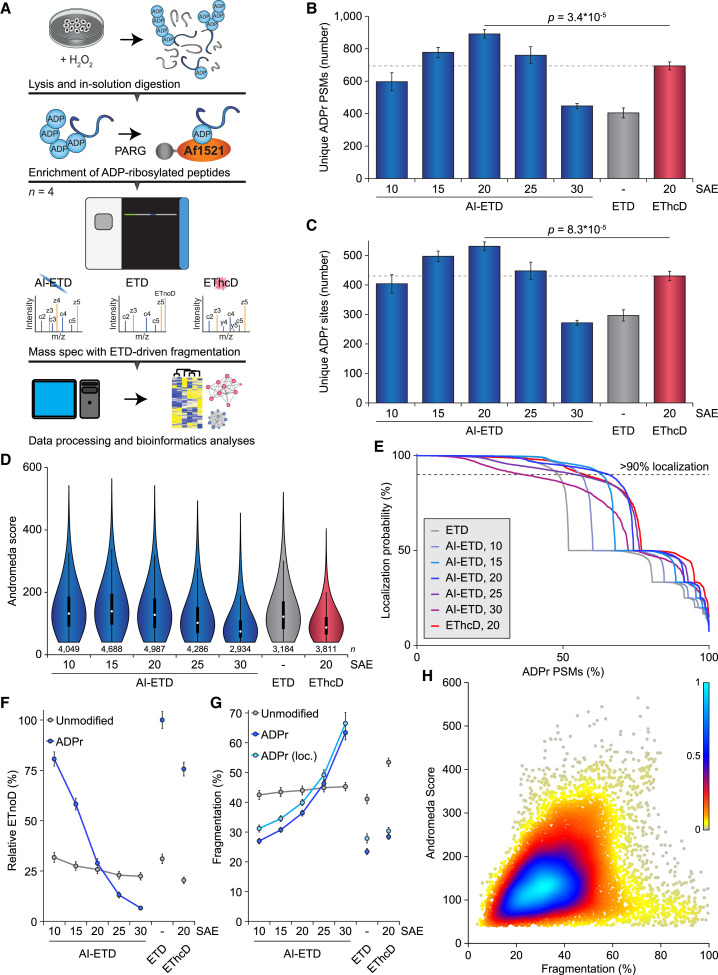
(A) Overview of the strategy employed to enrich ADPr peptides. All samples were analyzed in technical quadruplicate; n = 4.
(B) Overview of the number of ADPr peptide-spectrum matches (PSMs) identified and localized (>90% probability) for each dissociation method and supplemental activation energy (SAE; laser power for AI-ETD, normalized collision energy for EThcD). Significance was determined using two-tailed Student’s t testing. Error bars represent SD; n = 4 technical replicates.
(C) As in (B), but displaying the number of ADPr sites identified.
(D) As in (B), but displaying the spectral quality (in Andromeda score) of all identified ADPr-modified peptides. Distribution of data points is visualized: line limits, 1.5× interquartile range; box limits, 3rd and 1st quartiles; white dot, mean. Number of data points (n) is visualized below the distributions.
(E) ADP-ribose localization probability plotted against the ranked fraction of all PSMs. Note that although all probabilities are displayed, only those over 0.9 were used for assignment of ADPr peptides and sites.
(F) Visualization of the average relative degree of non-dissociative electron transfer (ETnoD). Derived from all peptide-identified MS/MS spectra, and separately visualized for unmodified and ADPr peptides. Error bars represent 5× SEM.
(G) Visualization of the average degree of precursor fragmentation, calculated by dividing observed fragment ion peak intensity by the sum of non-ETD, ETnoD, and all fragment ion peak intensities. Error bars represent 5× SEM.
(H) Spectral quality plotted against the average degree of precursor fragmentation for localized ADPr peptides. Coloring represents the relative density of dots in the plot, with higher values corresponding to higher density.