Figure 3.
Analysis of Physiological ADPr Using AI-ETD
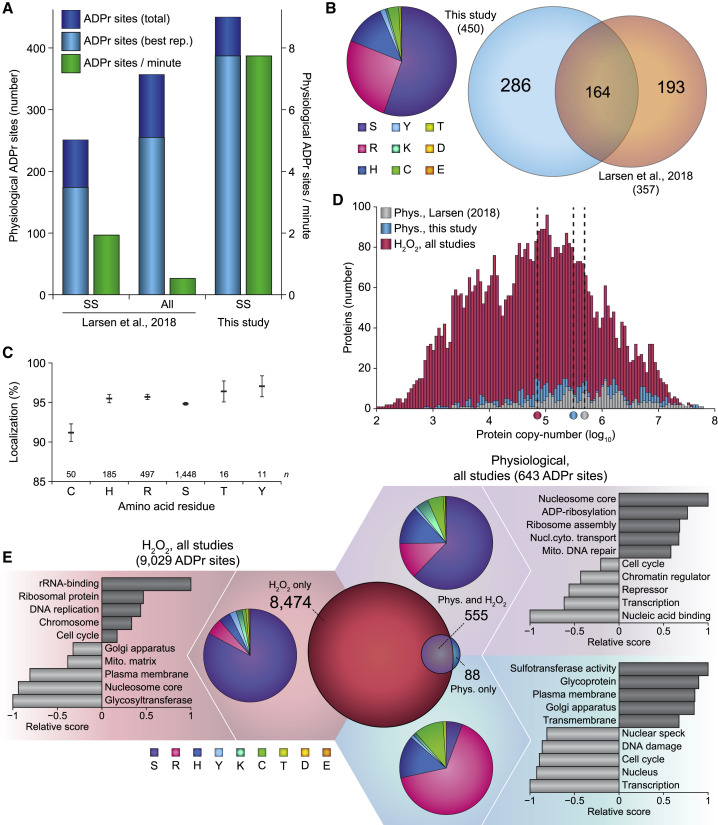
(A) Overview of the number of physiological ADPr sites identified in this study, compared with a previous study that also included physiological ADPr (Larsen et al., 2018). Total: unique ADPr sites identified across all four cell culture replicates; Best rep.: unique ADPr sites identified in the best replicate; SS: single-shot analysis; all: high-pH fractionated analysis.
(B) Left: pie chart overview of the numerical amino acid residue distribution of all physiological ADPr sites identified in this study. Right: scaled Venn diagram visualizing distribution of identified physiological ADPr sites in this study compared with our previous study (Larsen et al., 2018).
(C) Average localization probability across all ADPr PSMs that were at least partially localized (>51%) to each specific amino acid residue type. Only amino acids with >10 partially localized PSMs were included. Error bars represent SEM; number of data points (n) is indicated.
(D) Depth of sequencing analysis, plotting number of identified physiological ADPr target proteins versus their known copy numbers. Proteins ADP-ribosylated in response to H2O2 were derived from this study, as well as various other studies (Bilan et al., 2017; Jungmichel et al., 2013; Larsen et al., 2018; Martello et al., 2016; Zhang et al., 2013). Protein copy numbers were derived from a deep HeLa proteome study (Bekker-Jensen et al., 2017), which covers the vast majority (>99%) of all ADPr target proteins identified. Dotted lines with color indicator below represent the median protein copy number for ADPr target proteins within each respective subset.
(E) “Fidget spinner” analysis, composed of scaled Venn diagram (center) visualizing distribution of H2O2-induced and physiological ADPr sites; pie chart overviews (around the center) of the amino acid residue distribution of each subset of ADPr sites, as indicated by background coloring; term enrichment analyses (outermost graphs) using Gene Ontology annotations and UniProt keywords, comparing proteins identified to be ADP-ribosylated in each subset of ADPr sites as compared with the other subsets, indicated by background coloring. Relative score is based on multiplication of logarithms derived from the enrichment ratio and the q value. Terms were significant with q < 0.02, as determined by Fisher exact testing with Benjamini-Hochberg correction. The full-term enrichment analysis is available in Table S5B. Background and Venn coloring: red, H2O2 only; purple, both H2O2 and physiological; blue, physiological only.