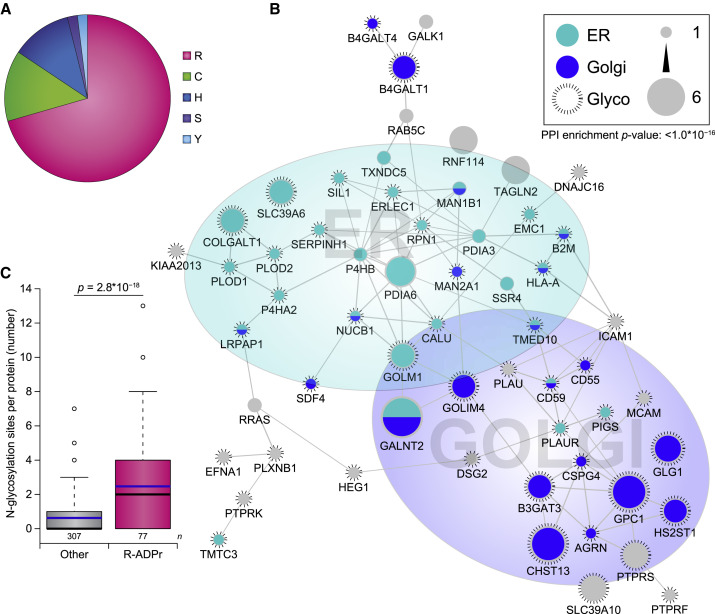
Figure 4.
Analysis of Arginine-Specific ADP-Ribosylation
(A) Pie chart of the amino acid residue distribution of ADPr target proteins exclusively modified under physiological conditions.
(B) STRING network visualizing functional interactions between proteins exclusively ADPr modified on arginine residues. Default STRING clustering confidence was used (p > 0.4), and disconnected proteins were omitted from the network unless they were identified by 2+ ADPr sites. Proteins were significantly interconnected, with a protein-protein interaction enrichment p value of 1.0 × 10−16.
(C) Distribution of the number of N-linked glycosylation sites per protein, as derived from a composite study (Sun et al., 2019). Line limit, 95th percentile; box limits, 3rd and 1st quartiles; black bar, median; blue bar, average. Number of data points (n) is visualized below the distributions. Significance was determined using two-tailed Student’s t testing.
ER, endoplasmic reticulum; Glyco, glycoprotein; Golgi, Golgi apparatus.