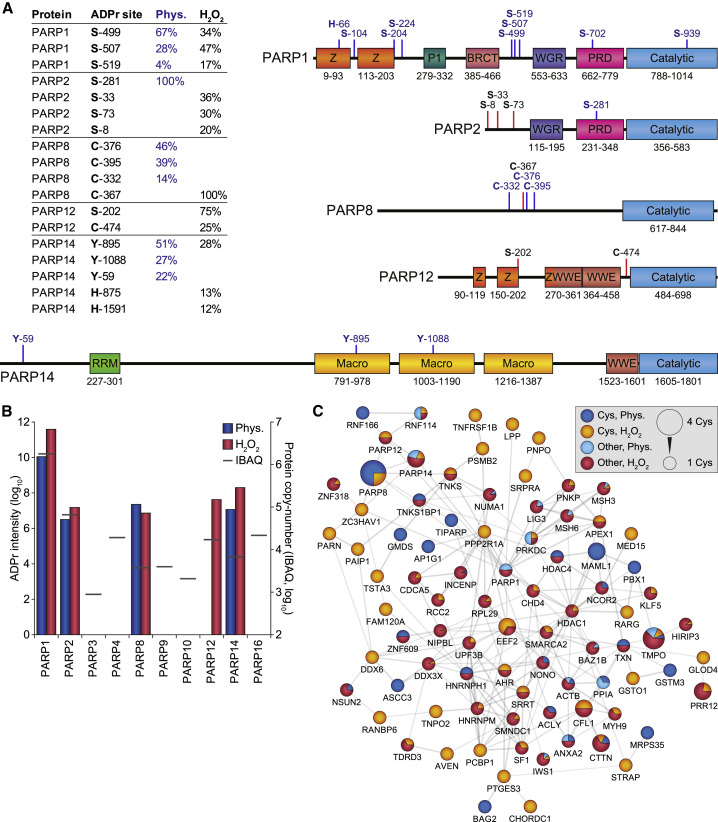
Figure 5.
ADP-Ribosylation of Different PARP Family Members
(A) Overview of the most abundantly ADPr-modified residues within PARP family members, as a fraction of total protein ADPr modification, either under physiological conditions (Phys.) or in response to H2O2. All physiologically detected ADPr sites, or otherwise up to four of the most abundant total ADPr sites, are indicated on the graphical representation of the PARP family members. Blue lines, physiological ADPr sites; red lines, H2O2-induced ADPr sites.
(B) Overview of the total amount of ADPr detected on each PARP family member, in relation to their known expression level (IBAQ). Both axes are logarithmic, and graph scaling was normalized to PARP1 and PARP2 values.
(C) STRING network visualizing functional interactions between proteins identified to be ADPr modified on cysteine residues in this study or in our previous study (Larsen et al., 2018). Default STRING clustering confidence was used (p > 0.4), and disconnected proteins were omitted from the network. Proteins were significantly interconnected, with a protein-protein interaction enrichment p value of 1.1 × 10−11. Distribution of color is relative to the number of sites within each category; sites detected both physiologically and in response to H2O2 were colored as physiological.
Cys, modification on cysteine residues; other, modification on non-cysteine residues.