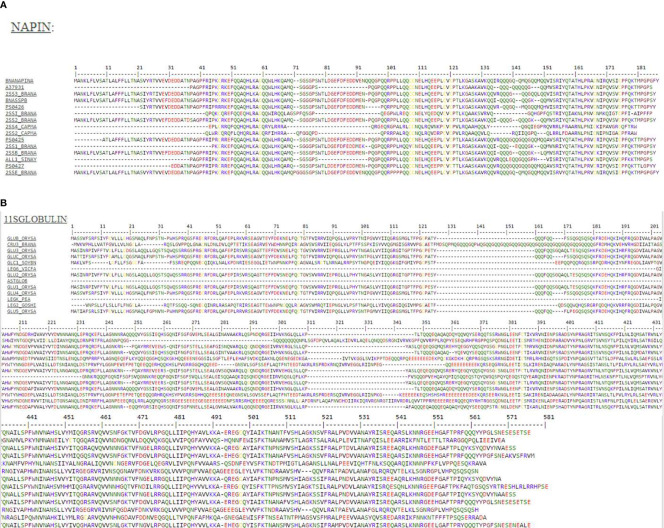
Figure 1.
(A) Protein motif analysis of antimicrobial signatures (shown in the same color) in napin genes using the PRINTS database. BNANAPINA = Q7DMU4_BRACM = Napin (Q7DMU4) from Brassica campestris; 2SS1_BRANA = Napin-1 (P01091), 2SSI_BRANA = Napin-1A (P24565), 2SSB_BRANA = Napin-B (P27740), 2SS2_BRANA = Napin-2 (P01090), 2SS3_BRANA = Napin-3 (P80208) and 2SSE_BRANA = Napin embryo-specific protein (P09893) are from Brassica napus, ALL1_SINAL = Allergen Sin a 1 (P09893) is from Sinapis alba, and 2SS2_CAPMA = Sweet protein mabinlin-2 (P30233) and 2SS4_CAPMA =Sweet protein mabinlin-4 (P80353) are from Capparis masaikai. The values in the parenthesis are the Uniprot accession numbers ( Supplementary Table 1 ). (B) Protein motif analysis of antimicrobial signatures (shown in the same color) in cruciferin genes using the PRINTS database. CRU3_BRANA = Cruciferin CRU1 (P33525) obtained from Brassica napus, GLUA1_ORYSJ = Glutelin type-A 1 (P07728), GLUA2_ORYSJ = Glutelin type-A 2 (P07730), GLUA3_ORYSJ = Glutelin type-A 3 (Q09151), GLUB1_ORYSJ = Glutelin type-B 1 (P14323) are obtained from Oryza sativa subsp. japonica; GLYG1_SOYBN = Glycinin G1 (P04776) from Glycine max, LEGA_GOSHI = Legumin A (P09802) from Gossypium hirsutum, LEGB6_VICFA = Legumin type B (P16079) from Vicia faba, LEGK_PEA = Legumin K (P05693) from Pisum sativum. The values in the parenthesis are the Uniprot accession numbers ( Supplementary Table 1 ).