Abstract
Myeloid cells co-expressing the markers CD11b, Ly-6G, and SiglecF can be found in large numbers in murine lung adenocarcinomas and accelerate cancer growth by fostering tumor cell invasion, angiogenesis, and immunosuppression; however, some of these cells’ fundamental features remain unexplored. Here, we show that tumor-infiltrating CD11b+ Ly-6G+ SiglecFhigh cells are bona fide mature neutrophils and therefore differ from other myeloid cells, including SiglecFhigh eosinophils, SiglecFhigh macrophages, and CD11b+ Ly-6G+ myeloid-derived suppressor cells. We further show that SiglecFhigh neutrophils gradually accumulate in growing tumors, where they can live for several days; this lifespan is in marked contrast to that of their SiglecFlow counterparts and neutrophils in general, which live for several hours only. Together, these findings reveal distinct attributes for tumor-promoting SiglecFhigh neutrophils and help explain their deleterious accumulation in the tumor bed.
Keywords: neutrophils, cancer, myeloid cells
Graphical Abstract
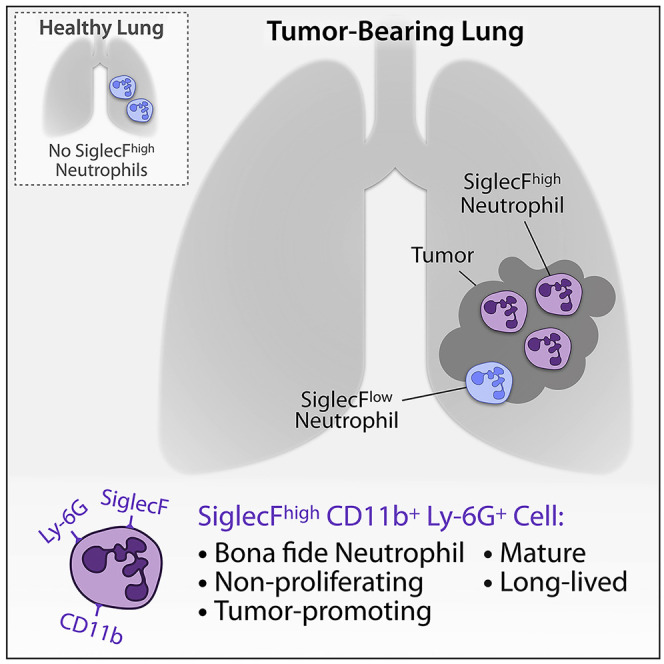
Neutrophils are essential defenders during inflammation and modulators of key cancer-associated activities with pro- or antitumor functions. However, cell markers that describe different neutrophil states are insufficiently known. Pfirschke et al. show that SiglecFhigh CD11b+ Ly-6G+ cells define mature and long-lived tumor-promoting neutrophils, which are distinct from other myeloid cells.
Introduction
Neutrophils are essential defenders against invading pathogens and orchestrators of tissue healing following injury. In the lung, these cells are important to deal effectively with air-bound pathogens. Produced in the bone marrow, neutrophils usually enter peripheral blood upon maturation and then home to inflamed tissues, where they can mediate potent effector functions, including internalization of pathogens for intracellular killing, release of proteases and reactive oxygen species, and formation of neutrophil extracellular traps. These activities can cause considerable collateral damage, effects that are most often limited in time in part because neutrophils are thought to be short lived (half-life of 6–8 h) and unable to divide (Ley et al., 2018; Ng et al., 2019; Summers et al., 2010).
Within tumors, high neutrophil counts are associated with poor clinical prognosis across many different cancer types (Gentles et al., 2015), suggesting that these cells, or at least some of them, broadly accelerate cancer progression (Engblom et al., 2016). Accordingly, experimental studies have shown that tumor-associated neutrophils, mostly defined as CD11b+ Ly-6G+ Ly-6Clow cells, can promote cancer outgrowth by awaking dormant tumor cells, fostering tumor cell proliferation, invasion, and metastasis; enhancing angiogenesis; and suppressing antitumor immunity (Engblom et al., 2017; Albrengues et al., 2018; Wellenstein et al., 2019; Szczerba et al., 2019), even though some tumor-associated neutrophil subsets can have antitumor effects (Ponzetta et al., 2019; Singhal et al., 2016; Fridlender et al., 2009; Matlung et al., 2018). Adding to this complexity, CD11b+ Ly-6Ghigh Ly-6Clow cells in cancer-bearing hosts are often broadly considered as immature cells with immunosuppressive functions and referred to as granulocytic myeloid-derived suppressor cells (G-MDSCs) (Veglia et al., 2018). The paucity of markers distinguishing neutrophil states from each other and from other myeloid populations has limited investigations of their diversity and pro- or antitumor functions.
Neutrophil-like CD11b+ Ly-6G+ Ly-6Clow cells in mouse lung adenocarcinomas include at least two subsets, which can be separated based on expression of the sialic-acid-binding protein SiglecF (Engblom et al., 2017). Whereas the SiglecFlow subset appears to be a “bystander” population in the tumor bed (it already exists in healthy lungs and neither promotes nor suppresses tumor progression), its SiglecFhigh counterpart instead shows phenotypes supporting tumor angiogenesis, tumor cell proliferation, extracellular matrix remodeling, and recruitment of immunosuppressive cells (Engblom et al., 2017). Accordingly, expression of a Siglecf high (but not a Siglecf low) gene signature in patients with lung adenocarcinoma is associated with worse disease outcome, suggesting that this signature may be a valuable biomarker of poorer prognosis (Engblom et al., 2017). A comparative scRNA-seq-based analysis of lung tumor-infiltrating immune cells in human and mouse further revealed conservation of several neutrophil states across these two species (Zilionis et al., 2019), indicating that studying neutrophils in mice should help to understand the human disease.
Here, we explored SiglecFhigh CD11b+ Ly-6G+ cells in mouse lung adenocarcinoma with the goal to further define these cells’ identity, compare them directly to previously described neutrophil and other myeloid cell populations, and understand the unfolding of the response mediated by these cells in vivo. Our findings reveal SiglecFhigh cells as bona fide neutrophils; yet these cells show fundamental features that diverge from those of previously described neutrophils and other myeloid cells, including G-MDSCs.
Results
SiglecFhigh CD11b+ Ly-6G+ Cells Resemble Neutrophils
To assess SiglecFhigh and SiglecFlow tumor-associated myeloid cells, we examined mice bearing KP1.9 lung adenocarcinomas that express G12D mutant oncogenic Kras and deleted alleles of the tumor suppressor gene Trp53 (Pfirschke et al., 2016). Lung adenocarcinoma nodules growing in these mice are infiltrated by myeloid cells, including monocytes, macrophages, and neutrophils (Cortez-Retamozo et al., 2012, 2013); this myeloid cell infiltrate is also observed in a sizable fraction of human lung adenocarcinomas (Lavin et al., 2017; Zilionis et al., 2019). Flow cytometry analysis of mouse lung tumor tissues identified several immune populations, including CD11b+ Ly-6G– SiglecFhigh cells, defined as eosinophils, and CD11b+ Ly-6G+ cells, defined as neutrophil-like cells (Figure 1 A). These two subsets showed distinct side-scatter and forward-scatter profiles as expected (Figures S1A and S1B). Around two-thirds of CD11b+ Ly-6G+ cells were SiglecFlow, as commonly reported for neutrophils, whereas the remaining one-third were SiglecFhigh (Figure 1A). The number of eosinophils, SiglecFlow neutrophils, and SiglecFhigh neutrophils, per milligram tissue, was greater in tumor-bearing compared to tumor-free lungs (Figure S1C).
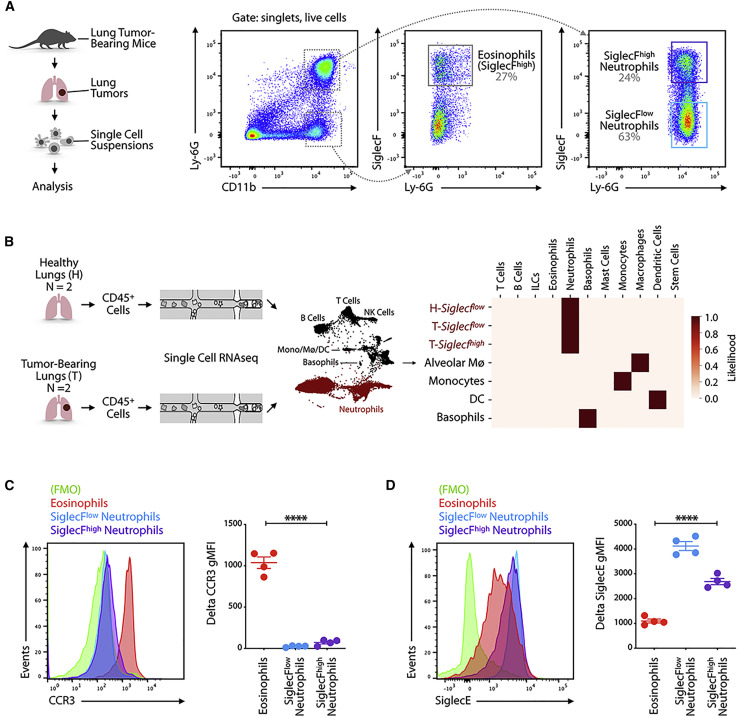
Figure 1.
SiglecFhigh CD11b+ Ly-6G+ Cells Resemble Neutrophils
(A) Cells obtained from lung tissue of KP1.9 tumor-bearing mice (day 29 after intravenous tumor cell injection) were stained by flow cytometry to identify eosinophils (CD11b+ Ly-6G− SiglecF+), SiglecFlow (CD11b+ Ly-6G+ SiglecFlow), and SiglecFhigh neutrophils (CD11b+ Ly-6G+ SiglecFhigh). Representative dot plots are shown (pre-gated on live cells).
(B) Suspensions of CD45+ cells for single-cell RNA sequencing were prepared from murine KP1.9 lung tumors (T) (n = 2) and lung tissue of healthy mice (H) (n = 2; Engblom et al., 2017; Zilionis et al., 2019). Major cell types (neutrophils highlighted in red) were identified by a Bayesian cell classifier as reported in Zilionis et al. (2019), and neutrophils were defined as tumor (T)-Siglecfhigh, tumor (T)-Siglecflow, and healthy (H)-Siglecflow based on the expression of genes correlated to SiglecF (Engblom et al., 2017). The heatmap shows a comparison of the 3 neutrophil subsets and of alveolar macrophages (Mø4), monocytes, dendritic cells (DCs), and basophils (rows) to immune profiles defined by the Immgen consortium (columns).
(C) Representative histogram (left) and quantification of geometric mean fluorescence intensity (gMFI) followed by fluorescence-minus one (FMO) signal subtraction (right) of CCR3 expression measured by flow cytometry in eosinophils and SiglecFlow and SiglecFhigh neutrophils (day 29 after intravenous tumor cell injection; n = 4 mice/group).
(D) Representative histogram (left) and quantification of delta gMFI (right) of SiglecE expression measured by flow cytometry in eosinophils and SiglecFlow and SiglecFhigh neutrophils (day 29 after intravenous tumor cell injection; n = 4 mice/group).
Data are represented as mean ± SEM. For comparisons between two groups, Student’s two-tailed t test was used. ∗∗∗∗p < 0.0001. See also Figure S1.
Considering that the marker SiglecF is frequently used to discriminate mouse eosinophils from neutrophils (Bochner, 2009; Zhang et al., 2004) and can also be expressed by lung macrophages (Engblom et al., 2017), we asked whether SiglecFhigh CD11b+ Ly-6G+ cells in lung tumors define bona fide neutrophils that express SiglecF or other immune cells, such as eosinophils, that upregulate Ly-6G. To this end, we first interrogated single-cell transcriptomic data of CD45+ cells from healthy lungs and KP1.9 lung tumors (Zilionis et al., 2019). We used a single-cell SiglecF expression score (Engblom et al., 2017) to operationally separate Siglecf low and Siglecf high cells resembling neutrophils in tumor tissue (T-Siglecf low and T-Siglecf high); we also assessed Siglecf low cells resembling neutrophils in healthy lungs (H-Siglecf low). We then annotated these populations with a Bayesian cell type classifier, which defines a set of labeled reference transcriptomes using publicly available bulk transcriptional profiles of annotated and sorted cell populations and returns a maximum likelihood cell type based on a multinomial model as well as the likelihoods themselves (Zemmour et al., 2018; Zilionis et al., 2019). This analysis revealed a maximum likelihood for T-Siglecf high cells to be neutrophils. The average likelihood for being a neutrophil or an eosinophil was ∼1 and 0, respectively (Figure 1B). As expected, both H-Siglecf low and T-Siglecf low cells also qualified as bona fide neutrophils (Figure 1B). Control myeloid cell types did not show a likelihood to be neutrophils; this included alveolar macrophages, which are referred to as Mø4 in Zilionis et al. (2019) and express SiglecF (Figure 1B). Of note, we could not identify eosinophils in our scRNA-seq datasets, even though the study was performed on total CD45+ cells. It is possible that eosinophils’ abundant RNase activity degrades mRNA before the reverse transcription reaction (Hämäläinen et al., 1999). Eosinophils are also absent from other scRNA-seq datasets (Azizi et al., 2018; Davidson et al., 2020; Zhang et al., 2020), further highlighting the difficulty to detect these cells by scRNA-seq.
We then used flow cytometry to measure the expression of cell surface protein markers that are commonly used to distinguish neutrophils from eosinophils. We found that SiglecFhigh CD11b+ Ly-6G− cells expressed the eosinophil marker CCR3 as expected (Höchstetter et al., 2000; Lee et al., 2012), whereas both SiglecFhigh and SiglecFlow CD11b+ Ly-6G+ cells were largely CCR3− (Figure 1C). Conversely, both SiglecFhigh and SiglecFlow CD11b+ Ly-6G+ cells expressed the neutrophil marker sialic-acid-binding protein E (SiglecE) (Zhang et al., 2004) at high levels, whereas SiglecFhigh CD11b+ Ly-6G− cells were SiglecElow (Figure 1D). Together, these data indicate that SiglecFhigh CD11b+ Ly-6G+ cells resemble neutrophils and not eosinophils.
SiglecFhigh Neutrophils Continuously Accumulate in Tumor-Bearing Lungs
SiglecFhigh neutrophils are absent from healthy lung tissue but highly abundant in KP1.9 tumor-bearing mice 1 month after tumor onset (Engblom et al., 2017). To gain insight into the unfolding of the tumor-driven SiglecFhigh neutrophil response, we investigated the abundance of these cells in lung tumor tissue at 5, 19, or 32 days after tumor onset (Figure 2 A). Lung weights at days 5 and 19 remained comparable to those of tumor-free mice, denoting limited tumor burden at these early time points, but then increased significantly at day 32 as expected (Engblom et al., 2017; Figure 2B). A small, yet detectable increase in abundance of SiglecFhigh neutrophils occurred as early as day 5 (Figures 2C and 2D). The abundance of SiglecFhigh neutrophils further increased on days 19 and 32, as tumors progressed (Figures 2C and 2D). By contrast, the abundance of SiglecFlow neutrophils in lung tumors remained largely unchanged during the same time frame (Figures 2C and 2D). Data for total CD11b+ Ly-6G+ neutrophils are shown in Figure S2A. The percentage of SiglecFhigh neutrophils in lung tumors strongly correlated with lung weight (p < 0.0001; r2 = 0.90; Figures 2E and S2B). Considering lung weight as a relevant proxy to lung tumor burden (Cortez-Retamozo et al., 2012), these data indicate “early” (e.g., at least on day 5) and continuous accumulation of tumor-associated SiglecFhigh neutrophils during cancer progression.
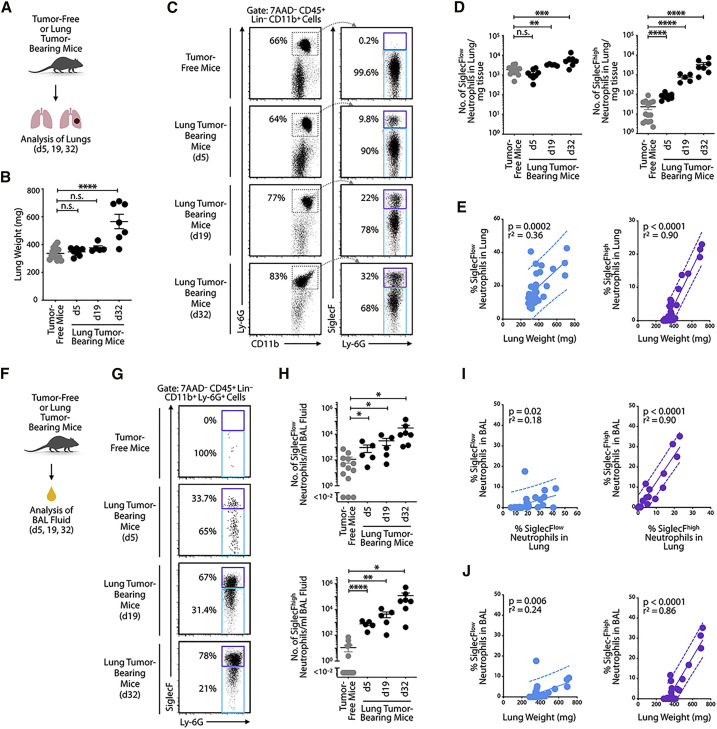
Figure 2.
SiglecFhigh Neutrophils Continuously Accumulate in Tumor-Bearing Lungs
(A) Diagram describing experimental procedure of the analysis of lung cells obtained from tumor-free or tumor-bearing mice.
(B) Lung weight (n = 5–14 mice/group) of tumor-free and KP1.9 tumor-bearing mice (days 5, 19, or 32 after intravenous tumor cell injection).
(C) Flow-cytometry-based detection of SiglecFhigh and SiglecFlow neutrophils from healthy lung tissue and KP1.9 lung tumors at different time points after tumor cell injection. Representative dot plots are shown (pre-gated on live CD45+ Lineage [Lin]− CD11b+ cells). The lineage master mix contained antibodies specific for CD90.2 and B220.
(D) Quantification of SiglecFlow (left) and SiglecFhigh (right) neutrophil numbers per mg lung tissue in tumor-free and tumor-bearing mice detected by flow cytometry (n = 5–14 mice/group).
(E) Lung SiglecFlow (left) and SiglecFhigh (right) neutrophils (percent of live cells measured by flow cytometry) plotted against lung weight (proxy of tumor burden) of KP1.9 lung-tumor-bearing or tumor-free mice (n = 34 mice) and linear regression was performed.
(F) Outline of experimental procedure for analysis of bronchoalveolar lavage (BAL) fluid-derived cells from tumor-free and KP1.9 lung cancer-bearing mice.
(G) Flow-cytometry-based detection of SiglecFhigh and SiglecFlow neutrophils from BAL fluid of healthy mice and mice carrying KP1.9 lung tumors at different time points after tumor cell injection. Representative dot plots are shown (pre-gated on live CD45+ Lin− CD11b+ Ly-6G+ cells). The lineage master mix contained antibodies specific for CD90.2 and B220.
(H) Quantification of SiglecFlow (top) and SiglecFhigh (bottom) neutrophils in BAL fluid of tumor-free and tumor-bearing mice detected by flow cytometry (n = 5–13 mice/group).
(I) Neutrophils in BAL fluid of KP1.9 lung-tumor-bearing or tumor-free mice (n = 30 mice) plotted against neutrophils in lung tissue, with performed linear regression. Data for SiglecFlow (left) and SiglecFhigh (right) neutrophils were measured by flow cytometry, and percent of live cells is shown.
(J) BAL fluid SiglecFlow (left) and SiglecFhigh (right) neutrophils (percent of live cells measured by flow cytometry) plotted against lung weight of KP1.9 lung-tumor-bearing or tumor-free mice (n = 30 mice) and linear regression was performed.
Data are represented as mean ± SEM. For comparisons between two groups, Student’s two-tailed t test was used. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. See also Figure S2.
Given that lung adenocarcinomas often occur in the peripheral regions of the airway epithelium, we investigated this space directly by collecting bronchoalveolar lavage (BAL) fluids (Figure 2F). This approach offers the possibility to sample the immune cell composition of peripheral airways and alveoli (Carvalho et al., 2017). Similar to our observations in whole-lung tissue, BAL fluid collected from tumor-free mice lacked SiglecFhigh neutrophils; however, some of these cells became detectable as early as day 5 after tumor onset and then gradually increased in number through days 19 and 32 (Figures 2G and 2H). The number of SiglecFlow neutrophils and total neutrophils in BAL fluid also increased, albeit at a slower pace (Figures 2G, 2H, and S2C). The percentage of SiglecFhigh neutrophils in BAL fluid correlated positively with the abundance of SiglecFhigh neutrophils in whole-lung tissue (p < 0.0001; r2 = 0.90; Figure 2I). Similarly, the percentage of SiglecFhigh neutrophils among live cells in BAL fluid correlated positively with lung weight (p < 0.0001; r2 = 0.86; Figures 2J and S2D). Figure S2E presents the correlation data for neutrophil numbers in lung tissue and BAL fluid. These data suggest that the tumor-associated SiglecFhigh neutrophil response detected in BAL fluid during tumor progression mirrors the one unfolding in whole-lung tumor tissue and is strongly and positively associated with lung tumor burden.
Siglec-Fhigh Neutrophils Are Mature, Non-proliferating Cells
Because immature neutrophils are often considered to promote cancer (Mackey et al., 2019; Veglia et al., 2018) and SiglecFhigh neutrophils exhibit potent tumor-promoting phenotypes and functions, including increased reactive oxygen species (ROS) production and ability to foster macrophage differentiation and to boost tumor cell proliferation in vivo (Engblom et al., 2017), we sought to assess the maturation status of these cells. Initially, we asked whether SiglecFhigh neutrophils can be found in the bone marrow, because this site produces neutrophils, many of which have not matured yet. However, we could not detect bone marrow SiglecFhigh neutrophils in lung tumor-bearing mice with high tumor burden (Figure S3A), suggesting that (1) SiglecFhigh neutrophils are phenotypically distinct from bone marrow neutrophils and (2) acquisition of SiglecF protein expression by neutrophils occurs after the cells are released from the medullary tissue. We also analyzed the spleen of KP1.9 lung-tumor-bearing mice for the presence of SiglecFhigh neutrophils, considering that this organ can sustain myelopoiesis in chronic inflammatory conditions, such as cancer (Swirski et al., 2009). However, we could not find SiglecFhigh neutrophils in the spleen (Figure S3B), suggesting that this organ does not produce these cells, even in the context of tumor-induced extramedullary myelopoiesis.
Because we also could not detect SiglecFhigh neutrophils in the peripheral blood of lung-tumor-bearing mice (Figure S3C), as we previously reported (Engblom et al., 2017), we further examined these cells in tumor tissue selectively. We assessed five parameters that are used to define neutrophil maturation, namely nuclear morphology, cell density, forward- and side-scatter profiles, cell surface protein expression, and cell proliferation. Neutrophils can be divided operationally into immature and mature cells, with the latter typically acquiring a segmented nucleus (including three to five lobes), a higher cell density, a modified cell phenotype, and inability to divide (Coffelt et al., 2016; Mackey et al., 2019; Ng et al., 2019).
First, to determine nuclear morphology, we purified SiglecFhigh and SiglecFlow neutrophils from lung tumors and analyzed them on cytospins upon hematoxylin and eosin staining. We found that the nuclei of nearly all SiglecFhigh neutrophils analyzed contained three or more lobes (Figure 3 A), which accords with the number of nuclear lobes of mature neutrophils (Chan et al., 2010; Veda, 2011). Also, the average number of nuclear lobes for SiglecFhigh neutrophils (3.50 ± 0.98; mean ± SD) aligned with that of their SiglecFlow counterparts (3.48 ± 0.98; mean ± SD), indicating that both neutrophil subsets are equally segmented and mature (Figure 3A).
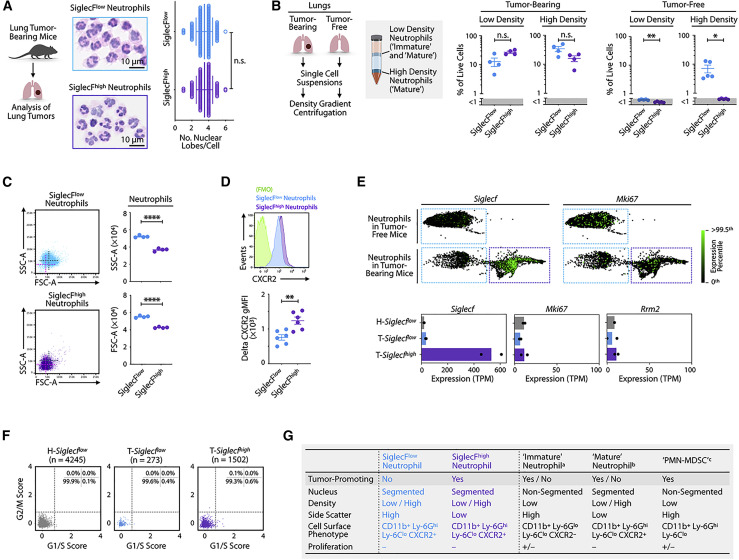
Figure 3.
SiglecFhigh Neutrophils are Mature, Non-proliferating Cells
(A) Quantification of nuclear lobes of fluorescence-activated cell sorting (FACS)-sorted SiglecFlow and SiglecFhigh neutrophils (30 cells/sample) from KP1.9 lung-cancer-bearing mice (day 29 after intravenous tumor cell injection; n = 4–5 mice/group) were investigated on hematoxylin and eosin (H&E)-stained cytospins. Representative cytospin images of SiglecFlow (top) and SiglecFhigh neutrophils (bottom) are presented. Scale bar, 10 μm.
(B) Flow-cytometry-based detection of SiglecFhigh and SiglecFlow neutrophils in low- and high-density cell layers obtained through Histopaque density gradient centrifugation of lung cells from tumor-bearing (n = 4 mice) and tumor-free mice (n = 15 mice; cells of three mice were pooled/sample). Neutrophils were analyzed on day 36 after intravenous KP1.9 tumor cell injection.
(C) Flow cytometry dot plots showing forward scatter (FSC) and side scatter (SSC) profiles of SiglecFhigh and SiglecFlow neutrophils obtained from KP1.9 tumor-bearing lungs (day 29 after intravenous tumor cell injection; n = 4 mice/group).
(D) Representative histogram (top) and quantification of geometric mean fluorescence intensity (gMFI) followed by fluorescence-minus one (FMO) signal subtraction (bottom) of CXCR2 expression measured by flow cytometry, in SiglecFlow and SiglecFhigh neutrophils (day 29 after intravenous tumor cell injection; n = 6 mice/group).
(E) (Top) Expression of the genes Siglecf and Mki67 in neutrophils from healthy and tumor-bearing mice. XY coordinates and expression values were obtained from Zilionis et al. (2019). The green color is saturated at the 99.5th expression percentile among neutrophils. Presentation of these data used SPRING, which is a tool for uncovering high-dimensional structure in single-cell gene expression data (Weinreb et al., 2018). Blue quadrants highlight SiglecFlow neutrophils and purple quadrants SiglecFhigh neutrophils. (Bottom) Average expression of selected genes in neutrophil subsets from KP1.9 lung tumors (T-Siglecflow and T-Siglecfhigh) or tumor-free lung (H-Siglecflow) is shown. Dots represent biological replicates.
(F) Relative expression of G1/S or G2/M gene signatures from Kowalczyk et al. (2015) and Tirosh et al. (2016) in T-Siglecflow, T-Siglecfhigh, and H-Siglecflow cells of two biological replicates. Expression values were obtained from Zilionis et al. (2019). Dashed lines indicate a relative expression of 0.8 that was set using Figure S3D as reference.
(G) Summary of cellular properties and functions of neutrophil subsets. The phenotypes of “immature” neutrophils (a), “mature” neutrophils (b), and “PMN-MDSCs” (c) are based on Chan et al. (2010), Eash et al. (2010), Evrard et al. (2018), Ley et al. (2018), Martin et al. (2003), Ng et al. (2019), Scapini et al. (2016), Veda (2011), and Veglia et al. (2018).
Data are represented as mean ± SEM or mean ± SD. For comparisons between two groups, Student’s two-tailed t test was used. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001. See also Figure S3.
Second, to define the density of SiglecFhigh neutrophils, we centrifuged single-cell suspensions of tumor-bearing lungs over a density gradient (Histopaque-1077 layered over Histopaque-1119; Swamydas et al., 2015) and then collected cells from so-called “low-density” (interphase of the plasma and Histopaque-1077 layers) and “high-density” (interphase of Histopaque-1077 and Histopaque-1119 layers) layers and further analyzed them by flow cytometry (Figure 3B). In steady state, mature and immature neutrophils are typically viewed as high- and low-density cells, respectively; however, mature neutrophils can show further heterogeneity in diseased hosts by including both high- and low-density subsets (Ley et al., 2018; Ng et al., 2019; Scapini et al., 2016). SiglecFhigh neutrophils from tumor-bearing lungs sedimented in both the low- and high-density layers (Figure 3B), indicating heterogenous density for these cells. We observed a similar distribution for SiglecFlow neutrophils and no significant enrichment of either neutrophil subset in the low- or high-density layers. Because both SiglecFlow and SiglecFhigh neutrophil subsets contain low- and high-density cell populations in tumor-bearing lungs, it is possible that both are heterogeneous. In control experiments, SiglecFlow neutrophils obtained from tumor-free lungs sedimented in the high-density layer exclusively, whereas SiglecFhigh neutrophils remained largely undetectable (Figure 3B), as expected.
Third, to identify the forward and side scatter of SiglecFhigh neutrophils, we used flow cytometry analysis of single-cell suspensions obtained from tumor-bearing lungs. We found a decrease in both variables for these cells, when compared to SiglecFlow neutrophils (Figure 3C).
Fourth, we also used flow cytometry to assess whether SiglecFhigh neutrophils express the chemokine receptor CXCR2, which is found on mature neutrophils (Eash et al., 2010; Martin et al., 2003). Tumor-infiltrating SiglecFhigh neutrophils expressed CXCR2 at even higher levels than their SiglecFlow counterparts (Figure 3D).
Fifth, to establish whether SiglecFhigh neutrophils include proliferating cells, we initially analyzed expression of two genes that are commonly associated with the cell cycle, namely Mki67 and Rrm2. Using single-cell transcriptomic data (Engblom et al., 2017; Zilionis et al., 2019), we found that T-Siglecf low, T-Siglecf high, and H-Siglecf low cells expressed similarly low levels of both cell proliferation marker genes (Figure 3E). To study cell proliferation genes more broadly, we considered G1/S and G2/M gene signatures, which are expressed by proliferating cells and include a total of ∼100 genes (Kowalczyk et al., 2015; Macosko et al., 2015; Tirosh et al., 2016). Neither T-Siglecf low, T-Siglecf high, nor H-Siglecf low subsets exhibited those signatures (Figures 3F, S3D, and S3E), suggesting that none of the neutrophil subsets are proliferating.
Thus, SiglecFhigh neutrophils in KP1.9 tumors are segmented, heterogeneous in density, low in forward and side scatter, express CXCR2, and not proliferating (Figure 3G). Considering all of these parameters, SiglecFhigh neutrophils resemble mature cells.
SiglecFhigh Neutrophils Are Long-Lived Cells within Tumors
Histological analysis of KP1.9 tumor-bearing lungs indicated that cells with a Ly-6G+ and SiglecF+ phenotype accumulated within tumor lesions (Figure 4 A). Considering that this process may require a prolonged amount of time, we sought to interrogate whether tumor-infiltrating neutrophils live longer than is typically thought (Ley et al., 2018; Ng et al., 2019; Summers et al., 2010). To address this question, we used parabiosis to establish shared circulation between CD45.1 and CD45.2 mice, each bearing KP1.9 tumors. The mice were separated 1 month later, and mouse lungs and blood were collected at days 0, 1, 4, and 6 after separation (n = 5–6 parabiotic mice for each time point; Figure 4B). Lung weights from all mice were not only similar but also higher than those of tumor-free mice (Figure 4C), indicating that tumors in parabiosed mice developed as expected.
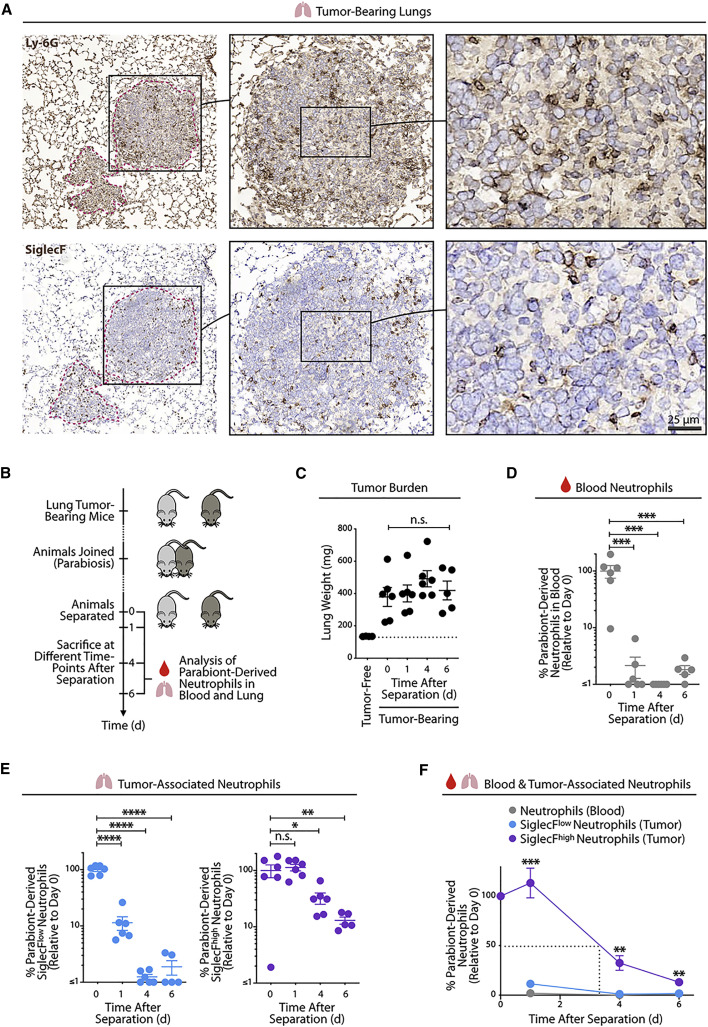
Figure 4.
SiglecFhigh Neutrophils Are Long-Lived Cells within Tumors
(A) Representative Ly-6G (top) and SiglecF (bottom) staining on cryopreserved KP1.9 lung tumor tissue sections of mice euthanized on day 29 after intravenous tumor cell injection. Tumor areas are highlighted by dotted pink lines. Scale bar, 25 μm.
(B) Diagram describing parabiosis and separation procedures on KP1.9 tumor-bearing mice (CD45.1 and CD45.2) to investigate the lifespan of parabiont-derived SiglecFhigh and SiglecFlow neutrophils in blood and lung tumors at different time points after separation (days 0, 1, 4, and 6). Mice were parabiosed 1 week after intravenous KP1.9 tumor-cell injection and separated on day 29.
(C) Lung weight of parabiosed tumor-bearing mice (n = 5–6 mice/time point) at time of euthanasia following separation. The lung weight of non-parabiosed tumor-free mice is shown as control (n = 4 mice).
(D) Quantification by flow cytometry of parabiont-derived CD11b+ Ly-6G+ neutrophils in blood of tumor-bearing mice (n = 5–6 mice/time point) at indicated post-separation time points.
(E) Flow-cytometry-based quantification of lung tumor-infiltrating SiglecFlow (left) and SiglecFhigh neutrophils (right) derived from the respective parabiont (n = 5–6 mice/time point).
(F) Percent of parabiont-derived neutrophils in blood and tumor-bearing lung after separation (n = 5–6 mice/time point). The dashed line indicates the half-life of SiglecFhigh neutrophils in tumor-bearing lung tissue.
Data are represented as mean ± SEM. For comparisons between two groups, Student’s two-tailed t test was used. For comparisons between three or more groups, one-way ANOVA with multiple comparisons was used. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Considering that surgical separation of two parabionts prevents further exchange of circulating cells between these mice, the survival of parabiont-derived cells in separated mice can be used as a direct measure of residence time in situ (Liu et al., 2007). Nearly all of the parabiont-derived neutrophils in peripheral blood were lost from each of the separated mice at day 1 already (the remaining cells accounted for only 1.8% ± 2.4% of those detected at day 0; mean ± SD; Figure 4D). These data align with the expected short lifespan of circulating neutrophils (Ley et al., 2018; Ng et al., 2019; Summers et al., 2010).
Analysis of lung tumor tissue revealed striking differences for SiglecFlow and SiglecFhigh neutrophils (Figure 4E). Parabiont-derived SiglecFlow neutrophils were largely lost at day 1 (the remaining cells accounted for only 11% ± 8% of those detected at day 0; mean ± SD) and disappeared at later time points (only 1.1% ± 0.4% and 1.6% ± 1.5% cells at days 4 and 6, respectively). By contrast, parabiont-derived SiglecFhigh neutrophils remained within tumors at day 1 (113% ± 37%) and continued to be detectable at days 4 (33% ± 18%) and 6 (13% ± 4%). In defining the half-life of cells as the duration until 50% of parabiont-derived cells are lost, it can be estimated that the half-life of SiglecFhigh neutrophils ranges between 3 and 4 days (Figure 4F). Our data do not permit a precise estimation of the half-life of SiglecFlow neutrophils, because these cells were nearly entirely lost at day 1 after separation; however, this half-life is likely to be lower than 1 day. Consequently, the half-life of tumor-infiltrating SiglecFhigh neutrophils is substantially longer than that of blood and lung SiglecFlow neutrophils in the same mice and of neutrophils in general.
Discussion
Neutrophils are increasingly recognized as cells that can promote cancer; however, we still have a limited understanding of their diversity and response to disease. Here, we centered on SiglecFhigh CD11b+ Ly-6G+ cells infiltrating murine lung adenocarcinomas and compared them to their SiglecFlow counterparts and other myeloid cell populations. Our findings indicate that SiglecFhigh CD11b+ Ly-6G+ cells can be viewed as a discrete population of mature neutrophils, which are molecularly, functionally, and/or morphologically distinct from SiglecFlow neutrophils, SiglecFhigh eosinophils, and CD11b+ Ly-6G+ G-MDSCs. In addition, we find that SiglecFhigh neutrophils can persist within tumors beyond the lifespan that is typically expected for neutrophils; this feature helps explain the deleterious accumulation of these cells within tumors.
First, our data indicate that SiglecFhigh CD11b+ Ly-6G+ cells are a discrete population of bona fide neutrophils. This finding is not immediately obvious, especially considering that (1) neutrophils typically do not express SiglecF, (2) SiglecF is a marker for the eosinophil lineage and alveolar macrophages in mice (Bochner, 2009), (3) some eosinophils may become Ly-6G+ (Percopo et al., 2017), and (4) neutrophils and eosinophils may resemble each other morphologically. Here, we found that tumor-infiltrating SiglecFhigh CD11b+ Ly-6G+ cells additionally expressed the neutrophil marker SiglecE, lacked the eosinophil marker CCR3, and more broadly classified fully as neutrophils, but not as eosinophils, based on their single-cell transcriptional profiles. These findings indicate that SiglecF alone may not be used to define cell lineages but rather that this protein can be expressed by various immune cell types, including eosinophils, some macrophages, and some neutrophils. The mechanisms regulating SiglecF expression in neutrophils and other cells in mice require further study.
Second, we find that SiglecFhigh neutrophils resemble mature and aged cells based on several parameters, including (1) a high level of nuclear segmentation, which increases throughout neutrophil maturation (Chan et al., 2010; Veda, 2011); (2) expression of CXCR2, which is required for the release of mature neutrophils from the bone marrow (Eash et al., 2010; Evrard et al., 2018; Martin et al., 2003); (3) lack of expression of proliferation markers and cell cycle gene signatures, which are expressed at high levels in developing neutrophils (Ng et al., 2019); and (4) absence of these cells in the bone marrow, a site primarily populated with immature neutrophils. These observations indicate that SiglecFhigh neutrophils are distinct from immature neutrophils or neutrophil-like cells, including G-MDSCs (Veglia et al., 2018), and align with previous indications that tumors can activate mature neutrophils in vivo (Scapini et al., 2016).
Human lung tumors may also contain mature neutrophils that resemble SiglecFhigh cells in mice. Interestingly, lung neutrophils form a continuum of states in both mice and humans (Zilionis et al., 2019). Mouse neutrophil states mN1,2 are highly enriched in healthy lungs, whereas mN3–6 expand in tumor-bearing lungs and include SiglecFhigh neutrophils (states mN4–6). SiglecF is not found in humans, but murine and human neutrophil states show conserved modules of gene expression, including neutrophil states that include SiglecFhigh cells in mice. Most notably, human neutrophil state hN5 resembles mN5, and both hN5 and mN5 define an end in the neutrophil state continuum (hN1→5; Zilionis et al., 2019). This suggests the existence in human tumors of mature neutrophils that resemble SiglecFhigh cells in mice. Also, the presence of hN5 is lung tumors is strongly associated with poor patient survival (Zilionis et al., 2019).
Third, our parabiosis studies suggest that SiglecFhigh neutrophils are long lived within tumors (half-life of ∼3–4 days), which is in marked contrast to the lifespan of neutrophils at steady state (half-life of ∼6–8 h only). This distinction is important because unchecked neutrophil activity can result in significant disease pathology (Amulic et al., 2012; Jorch and Kubes, 2017; Ley et al., 2018; Ng et al., 2019). Because SiglecFhigh neutrophils are absent from peripheral blood, parabiont-derived SiglecFhigh neutrophils detected at late time points after mouse separation may derive from SiglecFlow cells that subsequently acquired SiglecF expression. These cells could be bona fide SiglecFlow neutrophils or distinct precursors. Also, considering that SiglecFhigh neutrophils are homogeneously mature, it is unlikely that delayed (extramedullary) maturation of these cells would explain their extended lifespan within tumors. Instead, tumor-infiltrating SiglecFhigh neutrophils express genes that are associated with cell cycle arrest and reduced cell death (e.g., Cdkn1a and Cd47; Georgakilas et al., 2017; Jaiswal et al., 2009), which may help them resist cell death in ways that SiglecFlow neutrophils cannot.
The origin of SiglecFhigh neutrophils remains unknown. We previously found that ablation of osteoblasts selectively decreases the numbers of SiglecFhigh, but not SiglecFlow, CD11b+ Ly-6G+ cells in tumor-bearing lungs (Engblom et al., 2017), suggesting that these two neutrophil subsets originate from distinct bone marrow precursors. Also, we found increased expression of the Siglecf, Xbp1, and Ltc4s transcripts in circulating neutrophils from tumor-bearing mice, when compared to healthy mice (we could not find differences in protein expression). Considering that these transcripts are further upregulated in tumor-infiltrating SiglecFhigh neutrophils, these findings suggest that some circulating neutrophils may commit to a SiglecFhigh phenotype before seeding their destination tissue. However, it is formally possible that tumor-infiltrating SiglecFhigh neutrophils originate from bone fide SiglecFlow neutrophils that differentiate into SiglecFhigh cells upon reaching the tumor site and sensing local cues. This point may be addressed experimentally, at least in part, by tracking the fate of donor SiglecFlow neutrophils upon transfer into tumor-bearing recipient mice. We planned this experiment but could not complete it due to constraints imposed by the COVID-19 pandemic. At least, our data suggest that acquisition of SiglecF protein expression by neutrophils is a phenomenon that largely occurs upon arrival at the tumor site, because we could not detect SiglecFhigh cells in the bone marrow, spleen, and circulation.
Understanding the fate of SiglecFhigh neutrophils also requires study. Interestingly, SiglecFhigh neutrophils and senescent neutrophils (Casanova-Acebes et al., 2013) share common features, as they are both aged cells with hypersegmented nuclei and low forward- and side-scatter profiles. Yet these populations may also differ from each other, for example, based on Ly-6G and CXCR4 expression. Future studies should define whether SiglecFhigh neutrophils undergo senescence or apoptotic cell death in tumors, change phenotypes, and/or re-enter circulation (Wang et al., 2017).
The findings presented here provide fundamental insight into a neutrophil subtype that promotes cancer; they also bear importance for the development of immunotherapies aiming to target myeloid cells (Engblom et al., 2016; Weissleder and Pittet, 2020). For instance, finding ways to control SiglecFhigh neutrophils while sparing their SiglecFlow counterparts could be useful because these two populations have distinct functions. A better understanding of SiglecFhigh neutrophils should also have implications beyond cancer: since their discovery in 2017 in lung adenocarcinomas (Engblom et al., 2017), SiglecFhigh neutrophil-like cells have been reported in the ischemic heart after myocardial infarction (MI) (Vafadarnejad et al., 2019) and in the nasal mucosa in the context of allergic rhinitis (Matsui et al., 2020). The presence of these cells in various disease settings highlights the possibility that a SiglecFhigh neutrophil phenotype defines an inflammatory, rather than a tumor-specific, state. By extension, SiglecFhigh neutrophils may exhibit protective or deleterious functions, depending on the context in which they are found. A long-lived SiglecFhigh neutrophil may be detrimental in cancer but help fight some infections or promote tissue healing. Thus, it will be important to investigate SiglecFhigh neutrophils in the broad context of health and disease and uncover how these cells can be harnessed for therapy.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse CD45 (clone 30-F11) | Biolegend | Cat# 103126; RRID:AB_493535 |
| Anti-Mouse CCR3 (clone J073E5) | Biolegend | Cat# 144510; RRID:AB_2561609 |
| Anti-Mouse SiglecE (clone M1304A01) | Biolegend | Cat# 677106; RRID:AB_2566171 |
| Anti-Mouse CD45.2 (clone 104) | Biolegend | Cat# 109814; RRID:AB_389211 |
| Anti-Mouse CD45.2 (clone 104) | Biolegend | Cat# 109831; RRID:AB_10900256 |
| Anti-Mouse CXCR2 (clone SA044G4) | Biolegend | Cat# 149304; RRID:AB_2565692 |
| Anti-Mouse CD11b (clone M1/70) | BD Biosciences | Cat# 557657; RRID:AB_396772 |
| Anti-Mouse Ly-6G (clone1A8) | BD Biosciences | Cat# 551461; RRID:AB_394208 |
| Anti-Mouse Ly-6G (clone1A8) | BD Biosciences | Cat# 560599; RRID:AB_1727560 |
| Anti-Mouse Ly-6G (clone1A8) | Biolegend | Cat# 127618; RRID:AB_1877261 |
| Anti-Mouse Ly-6G (clone1A8) | Biolegend | Cat# 127602; RRID:AB_1089180 |
| Anti-Mouse Ly-6G (clone1A8) | Biolegend | Cat# 127643; RRID:AB_2565971 |
| Anti-Mouse SiglecF (clone E50-2440) | BD Biosciences | Cat# 564514; RRID:AB_2738833 |
| Anti-Mouse SiglecF (clone E50-2440) | BD Biosciences | Cat# 552125; RRID:AB_394340 |
| Anti-Mouse SiglecF (clone E50-2440) | BD Biosciences | Cat# 562681; RRID:AB_2722581 |
| Anti-Mouse SiglecF (clone E50-2440) | BD Biosciences | Cat# 565526; RRID:AB_2739281 |
| Anti-Mouse B220 (clone RA3-6B2) | BD Biosciences | Cat# 552772; RRID:AB_394458 |
| Anti-Mouse CD90.2 (clone 53-2.1) | BD Biosciences | Cat# 561642; RRID:AB_10895975 |
| Anti-Mouse CD45.1 (clone A20) | BD Biosciences | Cat# 560578; RRID:AB_1727488 |
| Anti-Mouse CD16/32 (clone 93), TruStain FcX | Biolegend | Cat# 101320; RRID:AB_1574975 |
| Biological Samples | ||
| Lungs from KP1.9 tumor-bearing or tumor-free mice | Mikael Pittet lab | N/A |
| Blood from KP1.9 tumor-bearing mice | Mikael Pittet lab | N/A |
| Spleens from KP1.9 tumor-bearing mice | Mikael Pittet lab | N/A |
| Bone marrow from KP1.9 tumor-bearing mice | Mikael Pittet lab | N/A |
| Bronchoalveolar lavage fluid from KP1.9 tumor-bearing or tumor-free mice | Mikael Pittet lab | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 7-aminoactinomycin D | Sigma | Cat# A9400 |
| Collagenase type I | Worthington | Cat# LS004197 |
| ACK lysis buffer | Lonza | Cat# 10-548E |
| Flow Cytometry Absolute Count Standard | Bangs Laboratories | Cat# 580 |
| Histopaque-1077 | Sigma-Aldrich | Cat# 10771 |
| Histopaque-1119 | Sigma-Aldrich | Cat# 11191 |
| Critical Commercial Assays | ||
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat# 554714 |
| Zombie Aqua Fixable Viability Kit | Biolegend | Cat# 423102 |
| ImmPRESS-AP Anti-Rat IgG, Mouse Adsorbed (alkaline phosphatase) Polymer Detection Kit | Vector Laboratories | Cat# MP-5444 |
| BLOXALL endogenous peroxidase blocking solution | Vector Laboratories | Cat# SP6000 |
| DAB Quanto Chromogen and Substrate | ThermoFisher Scientific | Cat# TA-060-QHDX |
| Deposited Data | ||
| Neutrophil single cell RNA sequencing data | (Engblom et al., 2017; Zilionis et al., 2019) | GEO: GSE127465 (Engblom et al., 2017; Zilionis et al., 2019) |
| Experimental Models: Cell Lines | ||
| Murine KP1.9 lung adenocarcinoma cell line derived from lung tumor nodules of a C57BL/6 KrasLSL-G12D/WT;Trp53flox/flox mouse | Alfred Zippelius | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J mice | Jackson Laboratory | Cat# 000664; RRID:IMSR_JAX:000664 |
| Mouse: B6.SJL-PtprcaPepcb/BoyJ | Jackson Laboratory | Cat# 002014; RRID:IMSR_JAX:002014 |
| Software and Algorithms | ||
| FlowJo v.8.8.7 and 10.4 | FlowJo, LLC | RRID:SCR_008520 |
| BD FACSDiva | BD Biosciences | RRID:SCR_001456 |
| GraphPad Prism v.8 | GraphPad Prism | RRID:SCR_002798 |
| Python 3.6.7 | Anaconda | https://www.anaconda.com/products/individual |
| NanoZoomer Digital Pathology (NDP.view2) | Hamamatsu | https://www.hamamatsu.com/eu/en/product/type/U12388-01/index.html |
| Aperio ImageScope | Leica | https://www.leicabiosystems.com/digital-pathology/manage/aperio-imagescope/ |
| Scanpy v. 1.4.4.post1 | (Wolf et al., 2018) | https://scanpy.readthedocs.io/en/1.4.4.post1/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mikael J. Pittet (mikael.pittet@unige.ch).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Single cell RNaseq data analyzed in this study is publicly available on GEO: GSE127465. Python code for selected analyses is provided as Methods S1: Python codes, related to STAR Methods.
Experimental Model and Subject Details
Cell lines
The lung adenocarcinoma cell line KP1.9 was derived from lung tumor nodules of a C57BL/6 male Kras LSL-G12D/WT; Trp53 flox/flox (KP) mouse and was kindly provided by Dr. Zippelius (University Hospital Basel, Switzerland). KP1.9 cells were maintained in Iscove’s DMEM cell culture media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin.
Mice
All mice used for this study were on a C57BL/6 background and male gender. Wild-type and CD45.1 mice were purchased from the Jackson Laboratory. All animals were housed under specific pathogen free conditions at the Massachusetts General Hospital (MGH). Animal experiments were approved by the MGH Institutional Animal Care and Use Committee (IACUC) and were performed in accordance with MGH IACUC regulations.
Mouse tumor model
Murine KP1.9 lung tumor cells were injected into 10-16 week old male mice intravenously (2.5x105 cells in 100μl phosphate buffered saline (PBS)) to develop orthotopic tumors in the lung. Mice were typically euthanized between 29-36 days after tumor cell injection. For some experiments, murine tissues were analyzed on earlier time-points. The exact time-point of analysis is indicated for the individual experiments in the respective figure legends. Tumor burden was scored by measuring postmortem lung weight as proxy for tumor burden as previously described (Pfirschke et al., 2016). Age and sex matched mice were used as tumor-free, healthy controls.
Method Details
Recovery of single cell suspensions from murine blood, lung, bone marrow and spleen
Single cell suspensions were isolated from murine blood, lung, bone marrow and spleen. The respective tissues and obtained single cell fractions were kept on ice or at 4°C for all steps if not stated otherwise.
Blood was collected into tubes containing 50 mM ethylenediaminetetraacetic acid (EDTA, Fisher Scientific) from the cheek or if mice were euthanized via cardiac puncture. Red blood cells were removed from Blood-EDTA mixtures using ammonium-chloride-potassium (ACK) lysis buffer (Lonza) for 5 min at room temperature, followed by a washing step with RPMI cell culture media. The resulting single-cell suspensions were washed and resuspended in flow cytometry washing buffer (PBS containing 0.5% bovine serum albumin (BSA, Fisher Scientific)).
Lungs were harvested, cut into small pieces using scissors and digested (RPMI media containing 0.2 mg/mL collagenase type I, Worthington Biochemical Corporation) for 1 h at 37°C while shaking (225 rpm). Femurs were harvested, cleaned and the bone marrow flushed out using a syringe containing cold flow cytometry washing buffer. Digested lung tissue, harvested bone marrow and harvested spleens were gently meshed through 40 μM cell strainers (Fisher Scientific) using a plunger. Red blood cells were removed from lung, bone marrow and spleen cells using 1 mL ACK lysis buffer for 1 min (lung, bone marrow) or 2 min (spleen) at room temperature and the reaction was stopped with RPMI media. The resulting single-cell suspensions were washed and resuspended in flow cytometry washing buffer until used for flow cytometry staining.
Recovery of single cell suspensions from murine bronchoalveolar lavage fluid
Bronchoalveolar lavage (BAL) fluid was removed from KP1.9 lung tumor-bearing and tumor-free mice to analyze neutrophil content. Mice were sacrificed, followed by the gentle removal of muscles around the neck to expose the trachea. In the next step, the ribs were cut to expose the heart as well as the lungs and a catheter (Exel International Disposable Safelet i.v. catheters, 22 G needle, Fisher Scientific) was carefully inserted into the trachea and tightened to the trachea using a nylon string to keep the catheter in place. A syringe (1 mL) containing 800 μl buffer (PBS containing 0.5% BSA) was connected to the catheter and the buffer content slowly injected and aspirated which caused the lung lobes to inflate and contract. This procedure was repeated two times before the syringe was removed from the catheter and the recovered fluid stored at 4°C. The flushing process was repeated for three times and all recovered BAL fluid pooled. Red blood cells were removed using 0.2 mL ACK lysis buffer for 1 min at room temperature and the reaction was stopped with RPMI media. Single-cell suspensions were washed and resuspended in flow cytometry washing buffer (PBS containing 0.5% BSA).
Flow cytometry
Single cell suspensions were obtained as described above. In some experiments, single cells were washed with PBS and dead cells stained using the Zombie Aqua fixable viability Kit (Biolegend) while incubating the cells for 20 min at room temperature. The cells were washed with flow cytometry staining buffer afterward (PBS containing 0.5% BSA and 2 mM EDTA) and incubated with FcBlock (TruStain FcX, anti-mouse CD16/32, clone 93, Biolegend) for 15 min followed by staining with fluorescent conjugated antibodies (Abs) for 45 min at 4°C. After a washing step using flow cytometry staining buffer, cells were fixed with the Fixation/Permeabilization Solution Kit (BD Biosciences) for 20 min at 4°C. The cells were washed with 1x perm wash buffer and analyzed on a LSRII flow cytometer (BD Biosciences) using BD FACSDiva and FlowJo software. In some experiments, 7-aminoactinomycin D (7AAD, Sigma) positivity was used to exclude dead cells if the Zombie Aqua fixable viability Kit was not used. In this case, single cells stained with fluorescent conjugated Abs were washed with flow cytometry staining buffer and incubated with 7AAD directly before analysis at the flow cytometer.
Doublet cells were excluded based on their forward/side scatter properties. The number of cells in a given tissue was calculated based on the percentage of each cell type identified by flow cytometry multiplied by the total number of cells in each organ. Cell numbers were obtained by determining viable cell numbers based on the trypan blue exclusion method or the Flow Cytometry Absolute Count Standard (Bangs Laboratories).
Based on cell surface marker expression following cell subsets were identified by flow cytometry: SiglecFlow neutrophils (CD45+ CD11b+ Ly-6G+ SiglecFlow), SiglecFhigh neutrophils (CD45+ CD11b+ Ly-6G+ SiglecFhigh), eosinophils (CD45+ CD11b+ Ly-6G— SiglecFhigh). In some experiments, a lineage Ab mix (of Abs specific for B220 and CD90.2) was used to remove lymphocytes. Following anti-mouse Abs were purchased from BD Biosciences: CD11b (clone M1/70), Ly-6G (clone 1A8), SiglecF (clone E50-2440), B220 (clone RA3-6B2), CD90.2 (clone 53-2.1), CD45.1 (clone A20); and Biolegend: CD45 (clone 30-F11), CCR3 (clone J073E5), SiglecE (clone M1304A01), CD45.2 (clone 104), CXCR2 (clone SA044G4).
Flow cytometry-based sorting of CD45+ cells from lung tissues
Single cell suspensions were obtained from lung tumor or healthy tumor-free lung tissue of age and sex matched C57BL/6 male mice which were perfused with PBS (37°C) as previously described (Engblom et al., 2017; Zilionis et al., 2019). Briefly, small equally sized tissue pieces of lung tumor tissue and tumor-free lungs were isolated using surgical fine scissors and digested (RPMI media containing 0.2 mg/mL collagenase type I) for 15 min at 37°C while shaking (700 rpm).
Digested lung tissue was gently meshed through 70 μM cell strainers using a plunger. Single cell suspensions were then stained with a fluorescent conjugated Ab specific to CD45 (clone 30-F11, Biolegend) for 45 min at 4°C. The cells were washed with flow cytometry staining buffer (PBS containing 0.5% BSA and 2 mM EDTA) and CD45+ live cells were sorted on a FACS Aria cell sorter (BD Biosciences) into FBS containing tubes which were kept on ice until the cells were further processed for single cell RNA sequencing (scRNA-seq). 7AAD was used to exclude dead cells.
Cytospin of neutrophils
SiglecFhigh (CD45+ CD11b+ Ly-6G+ SiglecFhigh) and SiglecFlow (CD45+ CD11b+ Ly-6G+ SiglecFlow) neutrophils were FACS sorted from lung tissue of KP1.9 tumor-bearing mice based on marker expression using the following anti-mouse mAbs: CD45 (clone 30-F11, Biolegend), Ly-6G (clone 1A8, Biolegend), CD11b (clone M1/70, BD) and SiglecF (clone E50-2440, BD Biosciences). 7AAD was used to exclude dead cells. Cytospins were performed using a Shandon Cytospin 4 centrifuge (Thermo Fisher Scientific).
In detail, 5x104 cells were centrifuged (500 rpm, 3 min) onto Tissue Path Superfrost Plus Gold microscope slides (Thermo Fisher Scientific) and dried overnight at room temperature. Cells were then fixed in 4% formaldehyde-buffered solution and stained with hematoxylin and eosin (H&E) using the Thermo Scientific Shandon Varistain Gemini ES Automated Slide Stainer. Image documentation was performed using the NanoZoomer 2.0-RS slide scanner system (Hamamatsu) or Leica Aperio AT2 slide scanner system and the NanoZoomer Digital Pathology and Aperio ImageScope software. 40x images were used to evaluate the number of nuclear lobes of sorted cells. Thirty cells per sample were analyzed. Two independent evaluations were performed and presented as number of nuclear lobes per cell.
Immunohistochemistry
For anti-SiglecF (rat anti-mouse Siglec-F mAb, clone E50-2440, BD Biosciences) and rat anti-mouse Ly-6G (clone 1A8, Biolegend) stainings, frozen tissue sections of murine lung tissue of KP1.9 tumor-bearing mice were generated, air-dried and fixed in acetone (−20°C) for 10 min. The sections were rehydrated and pretreated using BLOXALL endogenous enzyme blocking solution (Vector Laboratories) for 10 min to destroy all endogenous peroxidase activity. After blocking with normal goat serum, the sections were incubated with primary Abs for 1 h followed by several washes in PBS and secondary ImmPRESS polymer detection system (Vector Laboratories). DAB Quanto (Thermo Fisher Scientific) was applied as substrate and hematoxylin used as counterstain. For image documentation, the Leica Aperio AT2 slide scanner system or NanoZoomer 2.0-RS slide scanner system (Hamamatsu) with Aperio ImageScope or NanoZoomer Digital Pathology software were used.
Separation of neutrophils by density gradient centrifugation
Low density and high density neutrophils from lungs of KP1.9 tumor-bearing mice and healthy tumor-free mice were obtained through density gradient centrifugation. Lungs were harvested, cut into small pieces and digested with collagenase type I (0.2 mg/mL) in RPMI medium for 1 h at 37°C while shaking (225 rpm). Digested lung tissue was gently meshed through a 40 μM cell strainer using a plunger. Red blood cells were removed using 1 mL ACK lysis buffer (Lonza) for 1 min at room temperature, followed by a RPMI media washing step.
Cell pellets were resuspended in 1 mL PBS per tumor sample, while lung cells of three tumor-free mice were pooled. Histopaque gradients were generated in 15 mL reaction tubes by slowly overlaying 3 mL of Histopaque-1077 (density: 1.077 g/mL, Sigma-Aldrich) on 3 mL of Histopaque-1119 (density: 1.119 g/mL, Sigma-Aldrich). Cell suspensions were carefully added on top of Histopaque-1077, followed by a centrifugation step (30 min, 700 g, room temperature, without brake). Cells were collected at the interface of the Histopaque-1119 and the Histopaque-1077 layers (high density layer) and at the interface of the Histopaque-1077 and the plasma layers (low density layer). The collected cells were washed with flow cytometry staining buffer (PBS containing 0.5% BSA and 2 mM EDTA) and used for flow cytometry analyses to identify SiglecFhigh and SiglecFlow neutrophils.
Murine parabiosis and separation
Parabiosis and separation experiments were performed to study the longevity of SiglecFhigh and SiglecFlow neutrophils in lung tumor-bearing mice. One week post KP1.9 tumor cell injection (2.5x105 cells in 100μl PBS intravenously), lung tumor-bearing CD45.1 C57BL/6 male mice were parabiosed to lung tumor-bearing CD45.2 C57BL/6 male mice. The mice were separated on day 29 (d0) post tumor cell injection and neutrophils were analyzed at different time-points after separation (d0, d1, d4 and d6) in the blood and tumor-bearing lung. The experimental procedure was performed as previously described (Kamran et al., 2013).
Briefly, mice were anesthetized using an isoflurane vaporizer and shaved at corresponding lateral sides. Matching longitudinal skin incisions were made starting 0.5 cm above the elbow all the way to 0.5 cm below the knee joint and the skin was detached from the subcutaneous facia to create about 0.5 cm of free skin. The mice were joined by attaching the left olecranon of one parabiont to the right olecranon of the other parabiont using a non-absorbable 5.0 nylon suture (Ethilon), followed by the connection of the knee joints. After the attachment of the joints, the ventral and dorsal skin of the two animals were connected by a continuous suture.
For separation of the parabionts, mice were anesthetized, shaved at the area of the initial suture and separated through a longitudinal incision along the lateral suture. The newly formed fascia between the two mice was detached and the joints separated by cutting the knots of the suture connecting them. The skin of each separated mouse was reattached with a continuous suture. Blood chimerism was confirmed in all mice by analyzing blood drawn from the cheek three weeks after the joining procedure.
Identification of SiglecFhigh and SiglecFlow neutrophils in scRNA-seq data
Single cell gene expression data of SiglecFhigh and SiglecFlow neutrophils was obtained from Table S2 of Engblom et al. (2017). Data acquisition is briefly described here for completeness. InDrops scRNA-seq (Klein et al., 2015; Zilionis et al., 2017) was performed on FACS-sorted CD45+ cells from lungs of either tumor-free healthy (H; n = 2) or KP1.9 tumor-bearing (T; n = 2) mice. To identify single cell expression profiles corresponding to neutrophils among other CD45+ cells, a naive Bayesian classifier utilizing immune cell gene expression profiles from the Immgen consortium was used (Heng et al., 2008; described in detail in Zilionis et al., 2019). Fastq files, count matrices, cell type annotations, and xy coordinates from data visualization for all CD45+ cells are available on GEO: GSE127465 ( Zilionis et al., 2019). To further devide SiglecFhigh and SiglecFlow neutrophils (n = 6,020 cells), cells were classified based on a composite SiglecF expression score S. For each single cell k, we define: , where and is the percentile gene expression (dense ranking) of cell k for gene i, for the 50 most correlated genes to SiglecF (Spearman correlation), and is the corresponding sum of percentiles of the 50 most anticorrelated genes to SiglecF (gene lists are available in Table S3 of Engblom et al., 2017).
Quantification and Statistical Analysis
Comparison of immune cell expression profiles to published datasets
The heatmap in Figure 1B shows the likelihood of pseudo-bulk neutrophil expression profiles of 3 subsets we defined in Engblom et al. (2017) and additional immune cell types (alveolar macrophages (Mø4), monocytes, dendritic cells and basophils we defined in Zilionis et al. (2019) (rows) to be each of the immune populations (columns) defined by the Immunological Genome Project of the IMMGEN consortium (Heng et al., 2008). Likelihoods are obtained by applying a Bayesian classifier based on a multinomial model as described in detail in Zemmour et al. (2018) and Zilionis et al. (2019) and adapted from the approach by Jaitin et al. (2014). The code for recreating Figure 1B is provided as Methods S1: Python codes, related to STAR Methods.
G1/S and G2/M phase signature expression in immune populations
For each T-Siglecf low, T-Siglecf high, H-Siglecf low neutrophil, alveolar macrophage (Mø4), dendritic cell (DC1), B cell and proliferating T cell (T3) , a G1/S and G2/M phase signature score was calculated similar as in Tirosh et al. (2016) with the function sc.tl.score_genes_cell_cycle from the Scanpy package (Wolf et al., 2018). Briefly, given a gene list and a total cell count normalized, log transformed (i.e sc.pp.log1p), and scaled (i.e., sc.pp.scale) gene expression matrix (cell , gene ), a gene expression signature score for each cell was defined as: , where is a reference gene list that was generated by partitioning the vector of sum expression levels of all genes (i.e., , where belongs to ) into 25 bins, and then for each gene in selectingrandom genes from the same expression bin as that gene. To calculate the G1/S and G2/M phase signature scores for each T-Siglecf low, T-Siglecf high , H-Siglecf low neutrophil, Mø4, DC1, B cell and T3 T cell , G1/S and G2/M phase signature gene lists and were obtained from Table S5 of Tirosh et al. (2016) and and were calculated. The code for recreating Figures 3F, S3D, and S3E are provided as Methods S1: Python codes, related to STAR Methods.
Statistical analysis of flow cytometry and histology data
All statistical analyses were performed using GraphPad Prism software. Results were expressed as mean ± SEM or mean ± SD. Student’s two-tailed t test was done to compare two groups. One-way ANOVA with multiple comparisons was used to compare three or more groups. p values > 0.05 were considered not significant (n.s.); p values < 0.05 were considered significant. ∗ p value < 0.05, ∗∗ p value < 0.01, ∗∗∗ p value < 0.001, ∗∗∗∗ p value < 0.0001.
Acknowledgments
We thank the Harvard Stem Cell Institute for help with FACS sorting, the Single Cell Core Facility at Harvard Medical School for inDrop reagents, the Bauer Core Facility for sequencing, members of the Hope Babette Tang Histology Facility at the Koch Institute Swanson Biotechnology Center and Yoshiko Iwamoto from the MGH Center for Systems Biology for technical support, and Klein and Pittet lab members for helpful discussions. This work was supported in part by the Samana Cay MGH Research Scholar Fund, the Robert Wenner Award from the Swiss Cancer League, and NIH grants R01-AI084880, R01-CA218579, and R01-AI123349 (to M.J.P.); NIH grant R01-CA206890 (to M.J.P. and R.W.); and NIH grants R33-CA202064 and U01CA206997 (to R.W.). C.P. was supported in part by the MGH ECOR Tosteson Postdoctoral Fellowship; S.R. was supported by the MIT Ludwig Center for Molecular Oncology and funding from Richard O. Hynes (NIH grant U54-CA163109).
Author Contributions
C.P., C.E., and M.J.P. initiated the study, performed the analyses, and prepared the figures. C.P., C.E., J.G., Y.L., M.S., G.M.G., and A.K. performed mouse experiments. S.R. performed immunohistochemistry and analysis. R.Z. carried out scRNA-seq experiments. R.Z. and M.M. performed computational analyses. E.M., R.W., and A.M.K. provided input for research design and interpretation and edited the manuscript. M.J.P. supervised the study. M.J.P., C.P., and J.G. wrote the manuscript.
Declaration of Interests
M.J.P. has served as a consultant for Aileron Therapeutics, AstraZeneca, Cygnal Therapeutics, Elstar Therapeutics, KSQ Therapeutics, Merck, and Siamab Therapeutics. A.M.K. is a founder and shareholder in 1CellBio, Inc. These commercial relationships are unrelated to the current study. M.J.P., C.E., and C.P. are inventors on patent application 62/489,118 filed by MGH that covers the detection and targeting of tumor-promoting neutrophils.
Published: September 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108164.
Supporting Citations
The following references appear in the Supplemental Information: Hashimoto et al., 2013.
Supplemental Information
References
- Albrengues J., Shields M.A., Ng D., Park C.G., Ambrico A., Poindexter M.E., Upadhyay P., Uyeminami D.L., Pommier A., Küttner V. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018;361:1353. doi: 10.1126/science.aao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amulic B., Cazalet C., Hayes G.L., Metzler K.D., Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B.S. Siglec-8 on human eosinophils and mast cells, and Siglec-F on murine eosinophils, are functionally related inhibitory receptors. Clin. Exp. Allergy. 2009;39:317–324. doi: 10.1111/j.1365-2222.2008.03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A.S., Cuco C.M., Lavareda C., Miguel F., Ventura M., Almeida S., Pinto P., de Abreu T.T., Rodrigues L.V., Seixas S. Bronchoalveolar lavage proteomics in patients with suspected lung cancer. Sci. Rep. 2017;7:42190. doi: 10.1038/srep42190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Acebes M., Pitaval C., Weiss L.A., Nombela-Arrieta C., Chèvre R., A-González N., Kunisaki Y., Zhang D., van Rooijen N., Silberstein L.E. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell. 2013;153:1025–1035. doi: 10.1016/j.cell.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.-K., Tsai M.-H., Huang D.-C., Zheng Z.-H., Hung K.-D. Leukocyte nucleus segmentation and nucleus lobe counting. BMC Bioinformatics. 2010;11:558. doi: 10.1186/1471-2105-11-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt S.B., Wellenstein M.D., de Visser K.E. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- Cortez-Retamozo V., Etzrodt M., Newton A., Rauch P.J., Chudnovskiy A., Berger C., Ryan R.J., Iwamoto Y., Marinelli B., Gorbatov R. Origins of tumor-associated macrophages and neutrophils. Proc. Natl. Acad. Sci. USA. 2012;109:2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez-Retamozo V., Etzrodt M., Newton A., Ryan R., Pucci F., Sio S.W., Kuswanto W., Rauch P.J., Chudnovskiy A., Iwamoto Y. Angiotensin II drives the production of tumor-promoting macrophages. Immunity. 2013;38:296–308. doi: 10.1016/j.immuni.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S., Efremova M., Riedel A., Mahata B., Pramanik J., Huuhtanen J., Kar G., Vento-Tormo R., Hagai T., Chen X. Single-cell RNA sequencing reveals a dynamic stromal niche that supports tumor growth. Cell Rep. 2020;31:107628. doi: 10.1016/j.celrep.2020.107628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eash K.J., Greenbaum A.M., Gopalan P.K., Link D.C. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Invest. 2010;120:2423–2431. doi: 10.1172/JCI41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engblom C., Pfirschke C., Pittet M.J. The role of myeloid cells in cancer therapies. Nat. Rev. Cancer. 2016;16:447–462. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- Engblom C., Pfirschke C., Zilionis R., Da Silva Martins J., Bos S.A., Courties G., Rickelt S., Severe N., Baryawno N., Faget J. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science. 2017;358:eaal5081. doi: 10.1126/science.aal5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard M., Kwok I.W.H., Chong S.Z., Teng K.W.W., Becht E., Chen J., Sieow J.L., Penny H.L., Ching G.C., Devi S. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity. 2018;48:364–379.e8. doi: 10.1016/j.immuni.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Fridlender Z.G., Sun J., Kim S., Kapoor V., Cheng G., Ling L., Worthen G.S., Albelda S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakilas A.G., Martin O.A., Bonner W.M. p21: a two-faced genome guardian. Trends Mol. Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Hämäläinen M.M., Eskola J.U., Hellman J., Pulkki K. Major interference from leukocytes in reverse transcription-PCR identified as neurotoxin ribonuclease from eosinophils: detection of residual chronic myelogenous leukemia from cell lysates by use of an eosinophil-depleted cell preparation. Clin. Chem. 1999;45:465–471. [PubMed] [Google Scholar]
- Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S., Painter M.W., Immunological Genome Project Consortium The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Höchstetter R., Dobos G., Kimmig D., Dulkys Y., Kapp A., Elsner J. The CC chemokine receptor 3 CCR3 is functionally expressed on eosinophils but not on neutrophils. Eur. J. Immunol. 2000;30:2759–2764. doi: 10.1002/1521-4141(200010)30:10<2759::AID-IMMU2759>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Jaiswal S., Jamieson C.H.M., Pang W.W., Park C.Y., Chao M.P., Majeti R., Traver D., van Rooijen N., Weissman I.L. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A., Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorch S.K., Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 2017;23:279–287. doi: 10.1038/nm.4294. [DOI] [PubMed] [Google Scholar]
- Kamran P., Sereti K.I., Zhao P., Ali S.R., Weissman I.L., Ardehali R. Parabiosis in mice: a detailed protocol. J. Vis. Exp. 2013;80:50556. doi: 10.3791/50556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M.S., Tirosh I., Heckl D., Rao T.N., Dixit A., Haas B.J., Schneider R.K., Wagers A.J., Ebert B.L., Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.J., Jacobsen E.A., Ochkur S.I., McGarry M.P., Condjella R.M., Doyle A.D., Luo H., Zellner K.R., Protheroe C.A., Willetts L. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red”. J. Allergy Clin. Immunol. 2012;130:572–584. doi: 10.1016/j.jaci.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K., Hoffman H.M., Kubes P., Cassatella M.A., Zychlinsky A., Hedrick C.C., Catz S.D. Neutrophils: new insights and open questions. Sci. Immunol. 2018;3:eaat4579. doi: 10.1126/sciimmunol.aat4579. [DOI] [PubMed] [Google Scholar]
- Liu K., Waskow C., Liu X., Yao K., Hoh J., Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat. Immunol. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- Mackey J.B.G., Coffelt S.B., Carlin L.M. Neutrophil maturity in cancer. Front. Immunol. 2019;10:1912. doi: 10.3389/fimmu.2019.01912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Burdon P.C., Bridger G., Gutierrez-Ramos J.C., Williams T.J., Rankin S.M. Chemokines acting via CXCR2 and CXCR4 control the release of neutrophils from the bone marrow and their return following senescence. Immunity. 2003;19:583–593. doi: 10.1016/s1074-7613(03)00263-2. [DOI] [PubMed] [Google Scholar]
- Matlung H.L., Babes L., Zhao X.W., van Houdt M., Treffers L.W., van Rees D.J., Franke K., Schornagel K., Verkuijlen P., Janssen H. Neutrophils kill antibody-opsonized cancer cells by trogoptosis. Cell Rep. 2018;23:3946–3959.e6. doi: 10.1016/j.celrep.2018.05.082. [DOI] [PubMed] [Google Scholar]
- Matsui M., Nagakubo D., Satooka H., Hirata T. A novel Siglec-F+ neutrophil subset in the mouse nasal mucosa exhibits an activated phenotype and is increased in an allergic rhinitis model. Biochem. Biophys. Res. Commun. 2020;526:599–606. doi: 10.1016/j.bbrc.2020.03.122. [DOI] [PubMed] [Google Scholar]
- Ng L.G., Ostuni R., Hidalgo A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 2019;19:255–265. doi: 10.1038/s41577-019-0141-8. [DOI] [PubMed] [Google Scholar]
- Percopo C.M., Brenner T.A., Ma M., Kraemer L.S., Hakeem R.M., Lee J.J., Rosenberg H.F. SiglecF+Gr1hi eosinophils are a distinct subpopulation within the lungs of allergen-challenged mice. J. Leukoc. Biol. 2017;101:321–328. doi: 10.1189/jlb.3A0416-166R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfirschke C., Engblom C., Rickelt S., Cortez-Retamozo V., Garris C., Pucci F., Yamazaki T., Poirier-Colame V., Newton A., Redouane Y. Immunogenic chemotherapy sensitizes tumors to checkpoint blockade therapy. Immunity. 2016;44:343–354. doi: 10.1016/j.immuni.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetta A., Carriero R., Carnevale S., Barbagallo M., Molgora M., Perucchini C., Magrini E., Gianni F., Kunderfranco P., Polentarutti N. Neutrophils driving unconventional T cells mediate resistance against murine sarcomas and selected human tumors. Cell. 2019;178:346–360.e24. doi: 10.1016/j.cell.2019.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapini P., Marini O., Tecchio C., Cassatella M.A. Human neutrophils in the saga of cellular heterogeneity: insights and open questions. Immunol. Rev. 2016;273:48–60. doi: 10.1111/imr.12448. [DOI] [PubMed] [Google Scholar]
- Singhal S., Bhojnagarwala P.S., O’Brien S., Moon E.K., Garfall A.L., Rao A.S., Quatromoni J.G., Stephen T.L., Litzky L., Deshpande C. Origin and role of a subset of tumor-associated neutrophils with antigen-presenting cell features in early-stage human lung cancer. Cancer Cell. 2016;30:120–135. doi: 10.1016/j.ccell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers C., Rankin S.M., Condliffe A.M., Singh N., Peters A.M., Chilvers E.R. Neutrophil kinetics in health and disease. Trends Immunol. 2010;31:318–324. doi: 10.1016/j.it.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamydas M., Luo Y., Dorf M.E., Lionakis M.S. Isolation of mouse neutrophils. Curr. Protoc. Immunol. 2015;110:3.20.1–3.20.15. doi: 10.1002/0471142735.im0320s110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski F.K., Nahrendorf M., Etzrodt M., Wildgruber M., Cortez-Retamozo V., Panizzi P., Figueiredo J.L., Kohler R.H., Chudnovskiy A., Waterman P. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–616. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczerba B.M., Castro-Giner F., Vetter M., Krol I., Gkountela S., Landin J., Scheidmann M.C., Donato C., Scherrer R., Singer J. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafadarnejad E., Rizzo G., Krampert L., Arampatzi P., Nugroho V.A., Schulz D., Roesch M., Alayrac P., Vilar J., Silvestre J.-S. Time-resolved single-cell transcriptomics uncovers dynamics of cardiac neutrophil diversity in murine myocardial infarction. bioRxiv. 2019 doi: 10.1101/738005. [DOI] [Google Scholar]
- Veda P. Why are neutrophils polymorphonuclear? Eur. J. Inflamm. 2011;9:85–93. [Google Scholar]
- Veglia F., Perego M., Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hossain M., Thanabalasuriar A., Gunzer M., Meininger C., Kubes P. Visualizing the function and fate of neutrophils in sterile injury and repair. Science. 2017;358:111–116. doi: 10.1126/science.aam9690. [DOI] [PubMed] [Google Scholar]
- Weinreb C., Wolock S., Klein A.M. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics. 2018;34:1246–1248. doi: 10.1093/bioinformatics/btx792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R., Pittet M.J. The expanding landscape of inflammatory cells affecting cancer therapy. Nat. Biomed. Eng. 2020;4:489–498. doi: 10.1038/s41551-020-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenstein M.D., Coffelt S.B., Duits D.E.M., van Miltenburg M.H., Slagter M., de Rink I., Henneman L., Kas S.M., Prekovic S., Hau C.S. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature. 2019;572:538–542. doi: 10.1038/s41586-019-1450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D., Zilionis R., Kiner E., Klein A.M., Mathis D., Benoist C. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 2018;19:291–301. doi: 10.1038/s41590-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Q., Biedermann B., Nitschke L., Crocker P.R. The murine inhibitory receptor mSiglec-E is expressed broadly on cells of the innate immune system whereas mSiglec-F is restricted to eosinophils. Eur. J. Immunol. 2004;34:1175–1184. doi: 10.1002/eji.200324723. [DOI] [PubMed] [Google Scholar]
- Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A., He Y., Wang L., Zhang Q., Kim A. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459.e29. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- Zilionis R., Nainys J., Veres A., Savova V., Zemmour D., Klein A.M., Mazutis L. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 2017;12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D., Krishnan I., Maroni G., Meyerovitz C.V., Kerwin C.M. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNaseq data analyzed in this study is publicly available on GEO: GSE127465. Python code for selected analyses is provided as Methods S1: Python codes, related to STAR Methods.