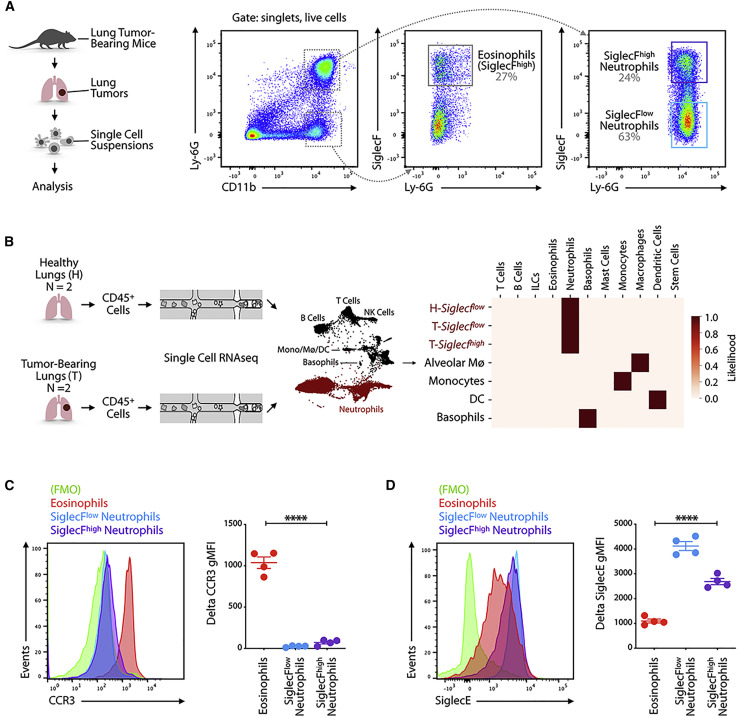
Figure 1.
SiglecFhigh CD11b+ Ly-6G+ Cells Resemble Neutrophils
(A) Cells obtained from lung tissue of KP1.9 tumor-bearing mice (day 29 after intravenous tumor cell injection) were stained by flow cytometry to identify eosinophils (CD11b+ Ly-6G− SiglecF+), SiglecFlow (CD11b+ Ly-6G+ SiglecFlow), and SiglecFhigh neutrophils (CD11b+ Ly-6G+ SiglecFhigh). Representative dot plots are shown (pre-gated on live cells).
(B) Suspensions of CD45+ cells for single-cell RNA sequencing were prepared from murine KP1.9 lung tumors (T) (n = 2) and lung tissue of healthy mice (H) (n = 2; Engblom et al., 2017; Zilionis et al., 2019). Major cell types (neutrophils highlighted in red) were identified by a Bayesian cell classifier as reported in Zilionis et al. (2019), and neutrophils were defined as tumor (T)-Siglecfhigh, tumor (T)-Siglecflow, and healthy (H)-Siglecflow based on the expression of genes correlated to SiglecF (Engblom et al., 2017). The heatmap shows a comparison of the 3 neutrophil subsets and of alveolar macrophages (Mø4), monocytes, dendritic cells (DCs), and basophils (rows) to immune profiles defined by the Immgen consortium (columns).
(C) Representative histogram (left) and quantification of geometric mean fluorescence intensity (gMFI) followed by fluorescence-minus one (FMO) signal subtraction (right) of CCR3 expression measured by flow cytometry in eosinophils and SiglecFlow and SiglecFhigh neutrophils (day 29 after intravenous tumor cell injection; n = 4 mice/group).
(D) Representative histogram (left) and quantification of delta gMFI (right) of SiglecE expression measured by flow cytometry in eosinophils and SiglecFlow and SiglecFhigh neutrophils (day 29 after intravenous tumor cell injection; n = 4 mice/group).
Data are represented as mean ± SEM. For comparisons between two groups, Student’s two-tailed t test was used. ∗∗∗∗p < 0.0001. See also Figure S1.