Table 2.
IC50s for inhibition of dimethylation activity and EC50s for abrogation of toxicity caused by GR15 or PR15 challenge, and chemical structures for each compound tested.
| Small molecule | IC50 (µM) ± s.e. | EC50 (µM) ± s.e. | Chemical structure | |||
|---|---|---|---|---|---|---|
| 3 µM GR15 challenge | 3 µM PR15 challenge | |||||
| WST-1 | LDH | WST-1 | LDH | |||
| MS023 | 0.545 ± 0.646 | 0.08 ± 0.02 | 0.012 ± 0.006 | 0.132 ± 0.017 | ~0.029 |
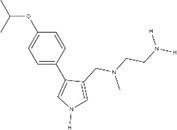
|
| MS049 | 1.082 ± 0.291 | ~2.103 | ~1.765 | ~1.99 | ~1.616 |
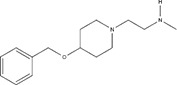
|
| EPZ020411 | 1.529 ± 0.310 | 12.01 ± 14.23 | 1.41 ± 0.118 | ~2.655 | ~2.939 |
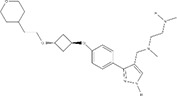
|
| GSK715 | 0.08 ± 0.441 | ~0.6724 | ~0.768 | ~0.799 | ~0.269 |
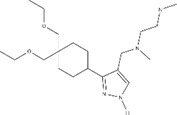
|
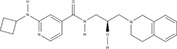
| GSK591* | 1.914 ± 96.28 | – | – | – | – |
|
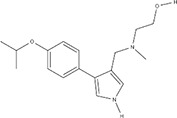
| MS094** | – | – | – | – | – |
|
IC50s were calculated using a four-parameter logistical regression model, and EC50s were calculated using a five-parameter logistical regression model. Ambiguous values result from curve fits that lack a plateau or constraint.
*Inhibits symmetrical dimethylation.
**Inert analog of MS023.
~indicates value is ambiguous.
