Figure 3: Specificity of uORF and N-terminal extension translation is influenced by condition and start codon identity.
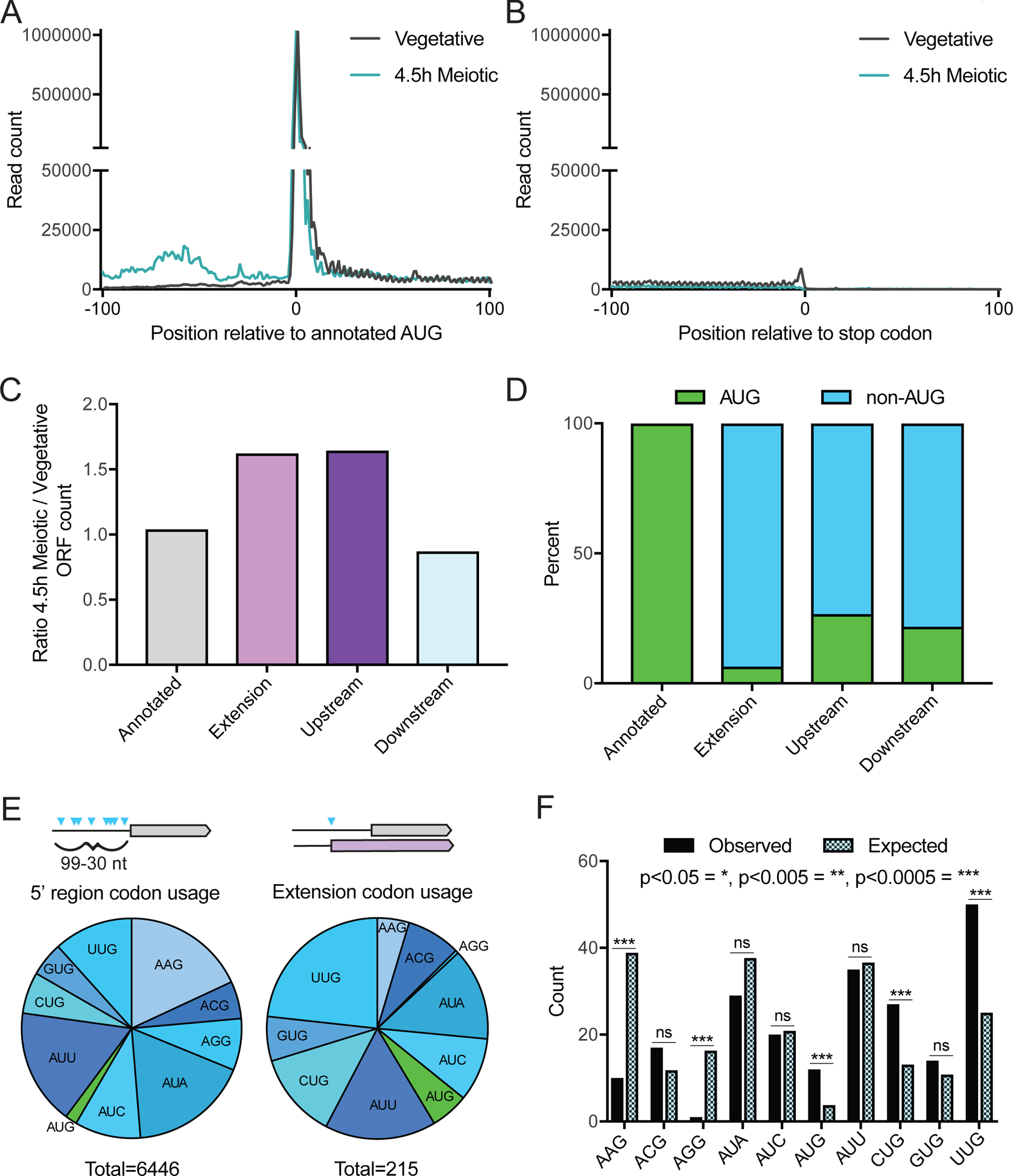
(A) Metagene plot of read counts from vegetative exponential and 4.5 hr time points, 100 nucleotides upstream and downstream of annotated AUG start codons. Reads are normalized to aligned reads for that timepoint. Increased read density is observed for the meiotic time point upstream of annotated start codons, but not after.
(B) Metagene plot of read counts from vegetative exponential and 4.5 h time points, 100 nucleotides upstream and downstream of annotated stop codons. Reads are normalized to aligned reads for that timepoint.
(C) Relative numbers of ORFs from different ORF categories, comparing the 4.5 h meiotic time point to vegetative exponential. More 5’ extensions and upstream ORFs are called in the meiotic time point, while annotated and downstream ORFs are similar between the two conditions.
(D) Percent of AUG versus non-AUG TISs for different ORF types. Annotated ORFs all have AUG start codons, while 5’ extensions, upstream, and downstream ORFs have primarily non-AUG TISs.
(E) Distribution of AUG and non-AUG start codon usage 99–30 nucleotides (nt) upstream of annotated AUG start codons for all possible TISs (left) and called 5’ extensions (right). Of the 6,446 sites possible in 5’ regions, 215 are observed to initiate translation of 5’ extensions called by ORF-RATER.
(F) Near-cognate codon usage for called extensions (observed) compared to relative abundance of all possible near-cognate codons within upstream regions (expected). Expected distribution is derived from counts of all possible TISs in the 99–30 nt upstream of annotated AUG start codons. P-values calculated by Fisher’s exact test, with p<0.05 = *, p<0.005 = **, p<0.0005 = ***, and ns = not significant.
