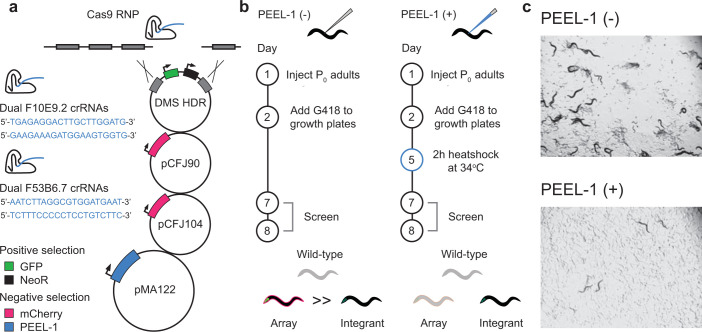
Fig 1. An optimized peel-1-DMS CRISPR-Cas9 genome editing pipeline kills arrays and spares genome-edited integrants.
(A) Schematic of the peel-1-DMS CRISPR-Cas9 genome editing method. Dual crRNAs targeting genes of interest are injected as RNPs in complex with Cas9 to induce double strand breaks. A homology-directed repair template is used to integrate a myo-2::GFP pharyngeal visual marker and a Neomycin resistance gene at the cut site for integrant positive selection. Co-injected extrachromosomal mCherry markers provide visual selection against arrays while peel-1 negative selection kills animals harboring arrays. In the standard DMS method arrays are manually distinguished from arrays based on dimness/consistency of GFP expression in the pharynx. (B) Injection and selection protocols/experimental design to test the efficacy of peel-1-DMS selection compared to our previously reported DMS method. (C) Peel-1-DMS selection kills arrays while sparing genome-edited integrants. Images from the F53B6.7 experiment 7 days post-injection.