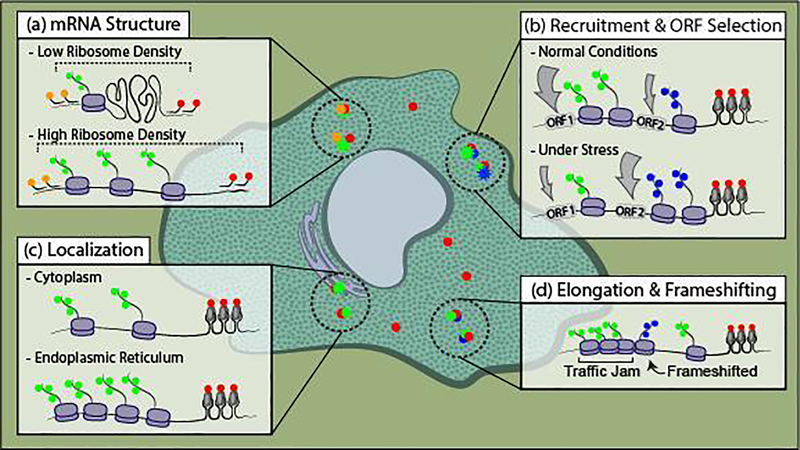
Figure 2. Recent applications of NCT to study active translation dynamics.
(a) NCT was combined with Single-Molecule Fluorescence In Situ Hybridization (smFISH) in fixed cells to measure the distance between the ends of the mRNA (red and orange). This was combined with antibody staining of nascent polypeptide chains (green) to count the translating ribosomes. By measuring the distance between 3’ (red) and 5’ (orange) mRNA probes and counting the number of translating ribosomes, 2D mRNA structures were quantified. (b) Two NCT systems were used to measure ribosome recruitment kinetics to a canonical (green) and non-canonical (blue) Open Reading Frame (ORF) and how ribosomes pick a start site once recruited to the mRNA. These assays were also conducted under stress conditions and differences in ribosome recruitment and ORF selection were measured. (c) By combining endoplasmic reticulum staining with NCT, mRNA localization and ribosomal content were determined. (d) NCT was used to investigate the heterogeneity of ribosomal elongation kinetics, including stalling at specific pause sites or frameshifting at a frameshift sequence. Both of which were associated with ribosomal traffic jams.