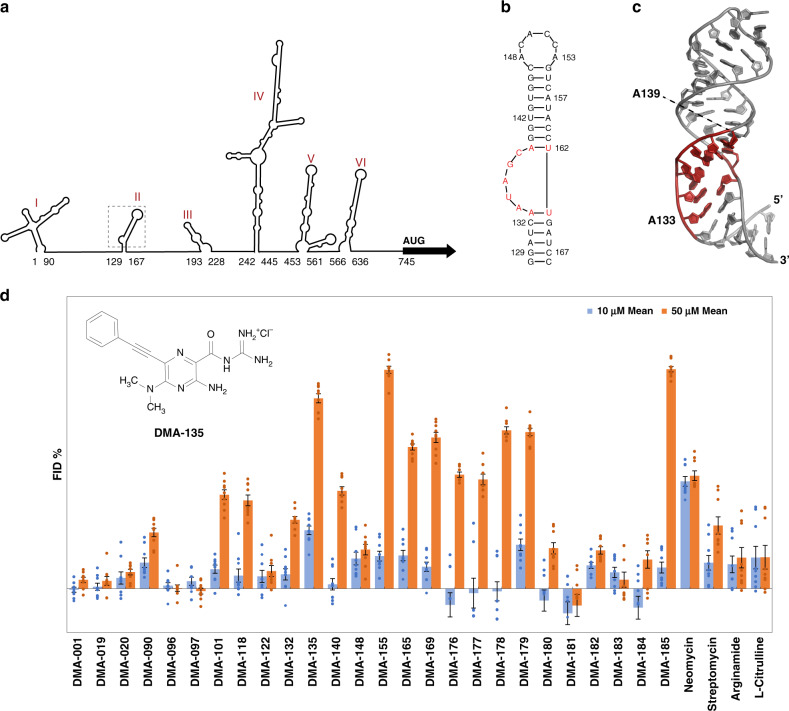
Fig. 1. RNA-biased small molecules bind to the EV71 SLII IRES domain.
a Schematic of the consensus secondary structure of the EV71 5′UTR determined using phylogenetic comparisons. b Secondary structure of EV71 SLII as validated by NMR spectroscopy9. c 3D structure (PDB code 5V16) of SLII refined against NOE-derived distant restraints and RDCs9. d Screening data for small molecules vs the EV71 SLII (wt) IRES domain at 10 and 50 µm concentrations. Error bars shown are standard errors of mean calculated from three individual replicates. Each replicate data point is the average of nine reads from three parallel readings on three wells (technical replicates). The chemical structure of DMA-135 is shown within the inset. Screening conditions were carried out in 50 mm Tris, 50 mm KCl, 0.01% Triton-X-100, 5% DMSO, pH = 7.4; Tat peptide: 60 nm; RNA: 90 nm; small molecule: 10 or 50 µm; excitation λ = 485 nm, emission λ = 590 nm; incubation time: 30 min.