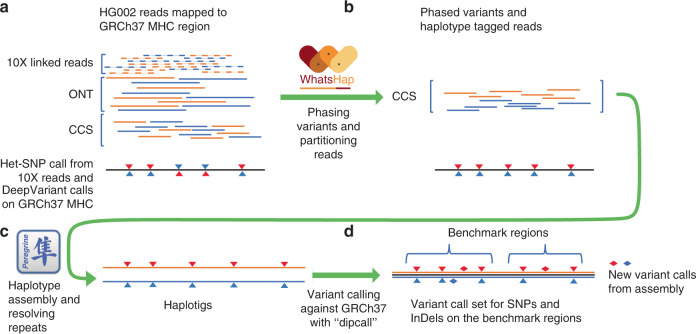
Fig. 1. Assembling a single contig for each haplotype.
a We regenotyped DeepVariant (DV) heterozygous SNVs with WhatsHap using Oxford Nanopore Technologies (ONT) and PacBio HiFi (CCS) reads to find a confident set of SNVs with concordant genotypes from DV/CCS, WhatsHap/ONT, and WhatsHap/CCS—our Confident HETs for phasing. We selected 10x Genomics (10X) variants with phased blocks from the 10X VCF. For phasing, we used WhatsHap to combine phased blocks from 10X with ONT reads to get a single phased block across the MHC. b We binned PacBio HiFi reads into two haplotypes, which are denoted as orange and blue reads, using WhatsHap. c We performed diploid assembly using the Peregrine Assembler with the haplotype-binned HiFi reads. d We generated the benchmark variant callset from the assembled haplotigs using dipcall, and defined benchmark regions excluding SVs, exceptionally divergent regions, low-quality regions in the assembly, and long homopolymers.