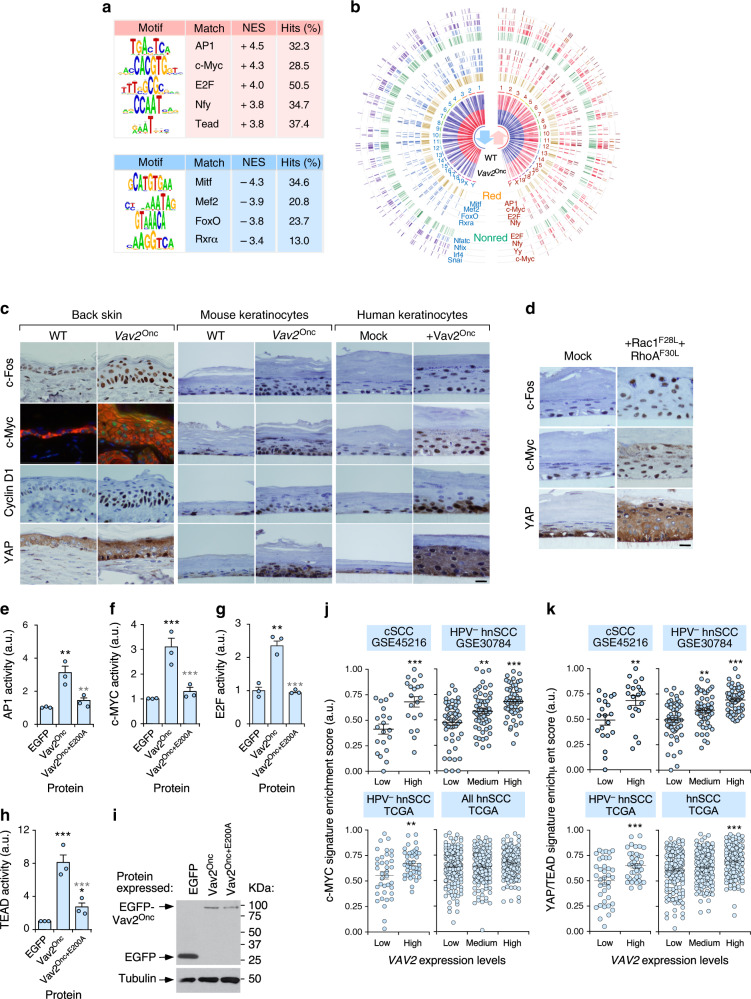
Fig. 6. Transcriptional factors involved in the Vav2Onc-dependent transcriptome.
a Transcription factor binding sites found enriched in the promoter regions of up- (red) and downregulated (blue) genes present in the “redundant” fraction of the Vav2Onc-driven transcriptome. The NES and percentage of hits for each transcription factor binding site are also indicated. b Circos plot of the differentially expressed genes in the Vav2Onc mouse skin. For each gene, the expression heatmap, the chromosomal location (1–19, X, Y), the binding sites for the indicated transcription factors, and the mirror-image behavior in the Vav2Onc-regulated transcriptome and the Vav2;Vav3-dependent fraction of the TPA-stimulated gene expression program in the skin of mice are indicated. Red, redundant; Nonred, nonreduntant. c Expression of indicated proteins (left) in the epidermis of the back skin of WT and Vav2Onc/Onc mice (left), 3D organotypic cultures from WT and Vav2Onc/Onc primary keratinocytes (middle), and 3D cultures of human keratinocytes (right). In the case of the immunofluorescence experiments using c-Myc antibodies (back skin, green color), sections were decorated with K14 (red color) and DAPI (blue color). Scale bar, 10 μm (n = 3 independent analyses for each experimental group). d Expression of indicated proteins (left) in 3D cultures of control and Rac1F28L + RhoAF30L-expressing human keratinocytes. Scale bar, 10 μm (n = 3 independent cultures). e–h Transiently transfected Vav2Onc triggers the rapid activation of endogenous AP1 (e), c-MYC (f), E2F- (g), and TEAD (h) proteins in human keratinocytes. Activity was measured using luciferase-encoding vectors containing promoter regions for each of the interrogated transcriptional factors, as described in Methods. Data represent the mean ± SEM. Black and gray stars indicate the P value of the indicated experimental values when compared to EGFP- and EGFP-Vav2Onc-transfected cells, respectively. *P = 0.011 (Vav2Onc+E200A) versus Vav2Onc, TEAD); **P = 0.008 (Vav2Onc versus EGFP, AP1), 0.010 (Vav2Onc+E200A) versus Vav2Onc, AP1), 0.002 (Vav2Onc versus EGFP, E2F); ***P < 0.0001 (all other tests) (ANOVA and Tukey’s HSD test, n = 3 independent experiments). i Expression of indicated proteins in one of the experiments performed in e to h. Tubulin was used as loading control (bottom panel) (n = 3 independent experiments). j, k Correlation between the levels of the VAV2 mRNA and the expression of c-MYC- (j) and YAP/TEAD-regulated- (k) gene signatures in the indicated (top) cSCC (n = 40) and hnSCC (n = 685) gene expression datasets. **P = 0.007 (VAV2 medium, MYC signature, GSE30784), 0.005 (VAV2 high, MYC signature, HPV- TCGA), 0.008 (VAV2 high, YAP signature, GSE45216), 0.002 (VAV2 medium, YAP signature, GSE30784); ***, P < 0.0001 (all other experiments) using the ANOVA and Dunnett’s multiple comparison test). Data represent the mean ± SEM. Source data for this figure are provided as a Source Data file.