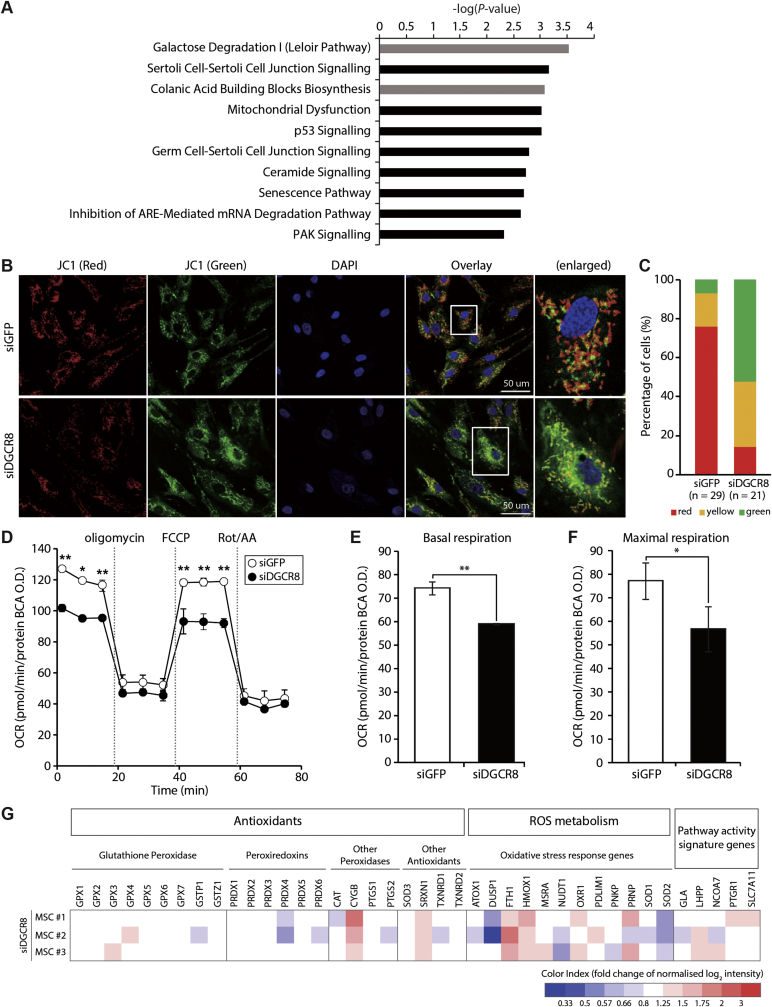
Fig. 3.
DGCR8 silencing induces mitochondrial dysfunction and deregulation of ROS metabolism-related genes. (A) Differentially expressed genes (DEGs) were subjected to Ingenuity Pathway Analysis (IPA), and the top 10 canonical pathways enriched in DGCR8-depleted hMSCs are shown. Metabolic pathways are shown in grey, while signalling pathways are indicated in black. (B) Mitochondrial membrane potential (MMP) in DGCR8-depleted hMSCs was assessed using the fluorescent dye JC-1. Representative confocal microscopy images showing high MMP (red fluorescence aggregates) and low MMP (green fluorescence monomers). Scale bars, 50 μm. (C) Quantification of JC-1 ratios. (D) Kinetic responses of oxygen consumption rate (OCR) in DGCR8-depleted hMSCs upon sequential addition of ATP synthase inhibitor oligomycin (1.5 μM), electron chain uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (1.0 μM), and complex I and III inhibitors rotenone and antimycin (1.5 μM each). The OCR values were normalised relative to the BCA protein assay. Error bars indicate the standard error of the mean of three independent experiments (*P < 0.05, **P < 0.01). (E) Basal mitochondrial respiration rates (basal OCR measurement minus rotenone/antimycin A response) and (F) maximal respiration rates (FCCP response minus rotenone/antimycin A response) were quantified in DGCR8 knockdown cells (*P < 0.05, **P < 0.01). (G) Heatmap showing the expression patterns of 40 oxidative stress-related genes upon DGCR8 depletion (n = 3 independent biological samples).