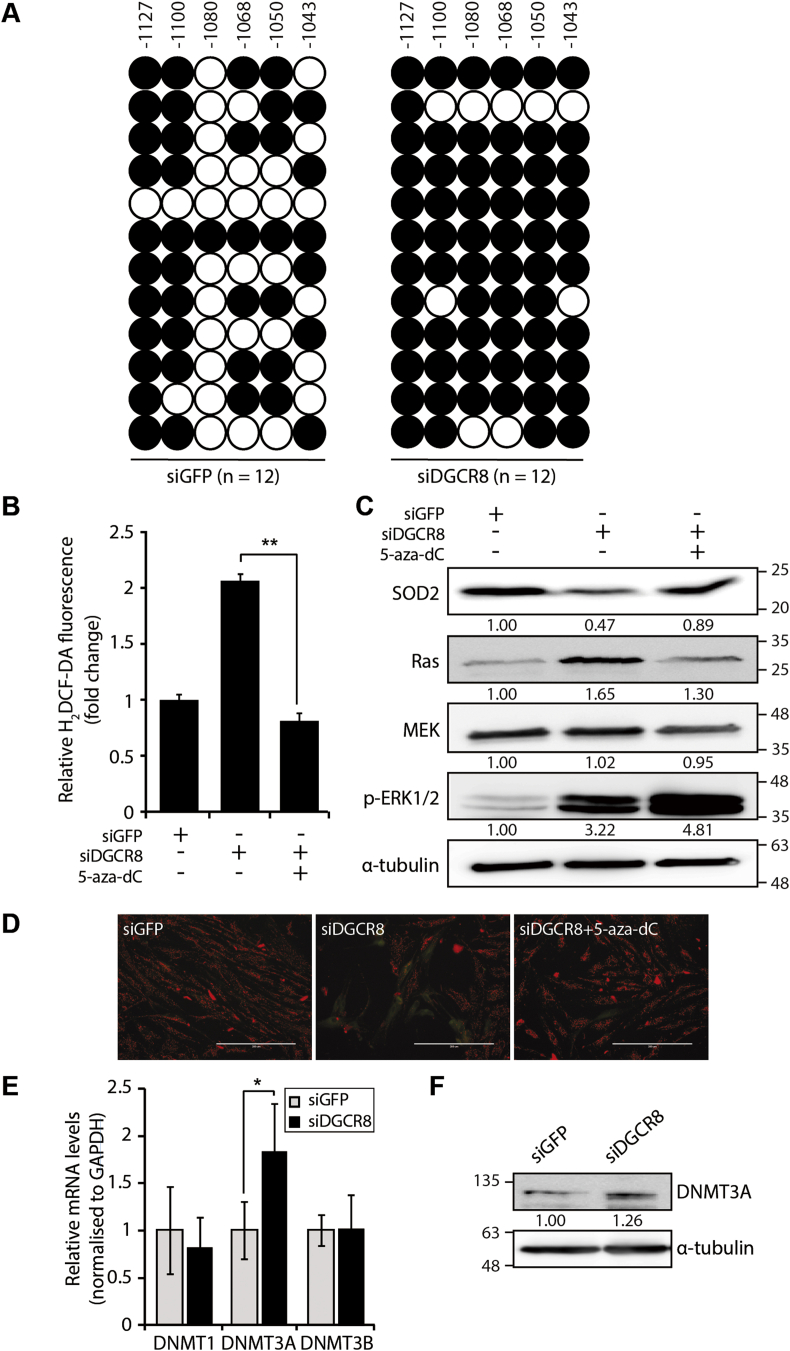
Fig. 5.
Hypermethylation of the SOD2 upstream regulatory region is dependent on DNMT3A. (A) Location and methylation status of six CpG sites in the SOD2 upstream regulatory region analysed by bisulphite sequencing (BSP) in control and DGCR8 knockdown hMSCs (n = 12 individual clones). Each circle represents a single sequencing reaction of each CpG site (white circles, unmethylated CpG sites; black circles, methylated CpGs). (B) Total ROS levels are measured by H2DCF-DA after 5-aza-dC treatment (2 μM) in DGCR8 knockdown hMSCs. Error bars denote the standard error of the mean of three independent experiments (**P < 0.01). (C) Immunoblot showing the levels of SOD2 and Ras signalling components after 5-aza-dC treatment (2 μM) in DGCR8 knockdown hMSCs. (D) Mitochondrial membrane potential (MMP) was assessed by JC-1 staining in DGCR8 knockdown hMSCs. Representative fluorescence microscopy images of siGFP- or siDGCR8-transfected cells, as well as 5-aza-dC-treated DGCR8 knockdown cells. Scale bars, 200 μm. (E) The mRNA levels of DNMTs were measured by quantitative real-time PCR. Data were normalised to GAPDH mRNA levels. Error bars indicate the standard error of the mean of three independent experiments (*P < 0.05). (F) Western blot analysis of DNMT3A protein levels in DGCR8 knockdown hMSCs.