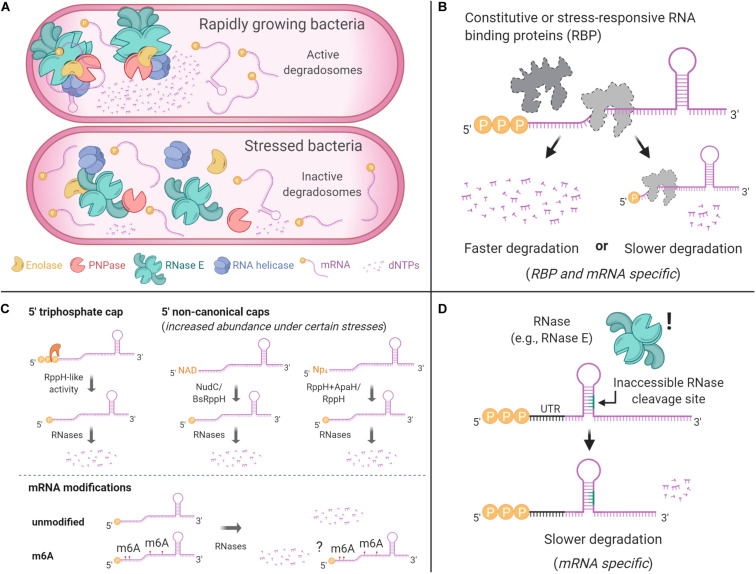
FIGURE 3.
Common mechanisms that can protect mRNAs from degradation. (A) Degradosome localization can influence its RNA degradation activity. In E. coli, the degradosome is anchored to the cytoplasmic membrane via RNase E’s N-terminal domain, where it displays higher RNA processing activity in degradation foci. A cytoplasmic RNase E is less efficient in degradosome assembly and RNA processing. In B. subtilis, RNase Y is associated with the membrane and is more active when in smaller foci and less active when in larger foci. (B) RNA binding proteins can modulate mRNA degradation. Some of them, such as CsrA in γ-Proteobacteria, have regulatory roles as a response to environmental changes. (C) The chemical nature of mRNA 5′ ends can protect transcripts from degradation. These caps may vary depending on stress conditions. Nucleotide modifications in the bodies of transcripts have also been reported, but they have not been shown to alter mRNA stability. (D) RNA degradation depends on RNase accessibility to cleavage sites. Secondary structures that block cleavage sites can result in slower RNA degradation.