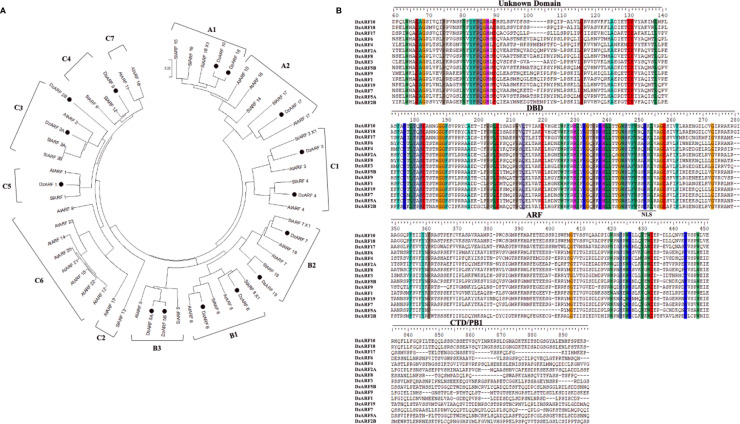
Figure 1.
Phylogenetic tree and multiple sequence alignment of the amino acid sequences of the DzARFs. (A) The deduced full-length protein sequences of durian ARFs (DzARFs) were aligned with Arabidopsis thaliana (AtARFs) and Solanum lycopersicum (SlARFs) protein sequences to construct the phylogenetic tree using MEGA X software and the neighbor-joining method (with 1,000 bootstrap replicates, a JTT model and pairwise gap deletion using a bootstrap test of phylogeny with the minimum evolution test and default parameters). (B) Multiple sequence alignment analysis was carried out using ClutalW. Identical amino acids are highlighted by color. Only important domains are presented (DBD, DNA binding domain; ARF, auxin response factor domain; CTD/PB1, carboxyl-terminal dimerization/Phox and Bem 1 domain). A nuclear localization signal (NLS) was identified at the end of DBD in all DzARFs.