Abstract
Background
Advanced sarcoma is a group of heterogeneous disease with poor prognosis and poor efficacy of medical treatment. They represent a promising group of tumors to assess molecular-based therapy (MBT) strategy.
Patients and methods
Genomic profiles of patients with advanced sarcoma included in the ProfiLER program were established by NGS using a 69 genes panel and CGH array. A weekly molecular board reviewed genomic reports to select relevant genomic alterations and propose recommendations for MBT.
Results
A genomic profile was available for 158 of 164 patients. At least 1 relevant genomic alteration was reported for 106 patients (67%), with frequent multiple alterations (68%). In total, 289 relevant genomic alterations were identified in 143 different genes; 139 homozygous deletions, 86 gene amplifications and 64 somatic mutations. The most frequently impacted genes were TP53, Rb1, CDKN2A, CDK4, MDM2, and PTEN. MBT was recommended for 47 patients and initiated for 13 patients.
One objective response was observed for an angiosarcoma treated with pazopanib for FLT4 amplification; 4 patients had a stable disease, including a long-lasting 33 months stabilization.
Conclusion
Genomic profiling for advanced sarcoma is feasible, even for bone sarcoma. A small proportion of patients are eventually treated with MBT, similar to other tumor types. We could not demonstrate this strategy to be beneficial to patients. Our data suggest that molecular profiling should not be used in routine practice but warrants further exploration in clinical trials, focusing on sarcoma with complex genomic, and adding transcriptomic analysis to the copy number and mutational analyses.
Keywords: Molecular profiling, Sarcoma, Precision medicine, Molecular-targeted therapy, Advanced cancer
Highlights
-
•
Rare and heterogeneous sarcoma represents a promising group of tumors to develop molecular-based therapy strategy.
-
•
Genomic profiling for advanced sarcoma is feasible. Relevant molecular alteration is identified in 67% of patients.
-
•
Few patients are eventually treated with molecular-based therapy, with no demonstrated benefit from this strategy.
-
•
Molecular profiling should not be used in routine practice but warrants further exploration in clinical trials.
Introduction
Sarcomas are a heterogeneous group of malignant tumors, gathering over 80 histological subtypes according to the latest WHO classification [1]. Sarcomas are rare diseases, with an overall annual incidence of 5.6 cases per 100,000 adults in Europe [2]. Sarcomas arise from mesenchymal cells from soft tissue (75%), 15% from organs (including gastrointestinal stromal tumor (GIST)) and 10% from bone tissue [[3], [4], [5]].
Sarcomas can be classified according to their genomic characteristics, driven by a single genomic abnormality (translocation, amplification, mutation or tumor suppressor gene loss) or harboring a complex genomic profile characterized by several gains, amplifications and losses of numerous chromosomes or chromosome regions, and numerous genetic translocations.
In the metastatic setting, patients with soft-tissue sarcoma (STS) have poor prognosis, with an overall survival (OS) ranging from 12 to 18 months and less than 20% patients are still alive at 2 years. Treatment of metastatic STS is mainly based on chemotherapy, the most efficient first-line treatment using doxorubicin, yielding to a tumor response rate of 10–14% [6].
The one-size-fits-all strategy developed so far in sarcoma demonstrated its limited efficacy because of the histological heterogeneity of the disease. Anthracycline-based chemotherapy failed to improve overall survival when administrated as adjuvant treatment or in metastatic setting in randomized studies including different histotypes. Better knowledge of biological mechanisms underlying carcinogenesis and disease progression in each subtype has become essential to identify reliable diagnosis, prognosis and, hopefully, predictive biomarkers [7]. The identification of genomic alterations occurring in the carcinogenic process has been used to provide new targeted therapies with the most convincing efficacy in the last decade. Tyrosine-kinase inhibitors targeting KIT or PDGFRA gain-of-function mutations in GIST are the best example of such reliable biomarkers [8]. Imatinib has also been used in dermatofibrosarcoma protuberans to target the COL1A1 and PGDFR rearrangement with impressive efficacy [9]. Large scale genomic screening has been implemented to better understand biology in sarcoma [[10], [11], [12], [13], [14], [15]]. Such programs are challenging and limited by low incidence of recurrent alterations; translation of genomic findings into clinical relevance remains complex despite proof of efficacy in preclinical studies, as MDM2 inhibition in liposarcoma for example.
Molecular based therapies have been proposed to target identified molecular alterations in each patient tumor. The predictive impact of a genomic alteration remains hypothetical in most of the cases. Whether the global population may benefit from genome-targeted therapy remains unclear [16]. Several studies such as MOSCATO [17] and SHIVA [18] trials showed limited efficacy and these series included only few sarcomas. However, such personalized therapeutic approach has to be explored, especially in rare and heterogeneous cancer like sarcoma.
The ProfiLER program is a multicentric open-label prospective trial establishing the genetic tumor profile of patients with any types of advanced cancer in order to propose molecular-based therapy [19]. The present study used data from ProfiLER to describe the frequency and distribution of actionable alterations in locally advanced or metastatic sarcomas, to explore their use for guiding molecular targeted-agents recommendations, and to describe responses to molecular-based recommended therapy.
Methods and materials
Study design and patients
The ProfiLER program aimed at establishing the genetic profile of patients with advanced cancer to guide recommendations of targeted therapies. The trial was conducted in four French authorized institutions, and coordinated by the Centre Léon Bérard (Lyon, France) after local approval (Ethics Committee of Lyon Sud-Est IV), in accordance with the Good Clinical Practice guidelines of the International Conference on Harmonization and the Declaration of Helsinki, and relevant French and European laws and directives. All patients signed a written informed consent before enrolment [19].
Patients were included in case of confirmed histological or cytological diagnosis of advanced (locally-advanced or metastatic) malignant tumor of any histological type, progressive after at least one prior line of standard treatment, with no access to curative treatment, available tumor sample -either formalin-fixed and paraffin embedded (FFPE) archived tumor sample or de novo biopsy for accessible lesion, left at the discretion of investigators. Patients for whom no tumor sample was available were excluded.
Clinical information (patient, tumor and treatment characteristics) were prospectively collected. To perform this analysis, patients diagnosed with soft-tissue sarcoma, GIST and bone sarcoma were selected, excluding carcinosarcoma because of their epithelial origin.
Procedures
FFPE tumor specimen containing ≥10% of tumor cells (as assessed by anatomopathologists on H&E slides) were used to determine genetic molecular profiles by Next Generation Sequencing (NGS) using two successive gene panels of 59 and 69 genes, respectively (Supplementary Table S1), and genome wide microarray-based Comparative Genomic Hybridization (aCGH). The samples with insufficient DNA quantity for standard aCGH were analyzed using Cytoscan HD (Affymetrix®, Santa Clara, CA, USA) according to manufacturer protocol.
The platform Ion Torrent PGM (Thermo Fisher Scientific, Massachusetts, USA) was used for mutation screening. Each tumor sample used 10 ng DNA for library construction using Ion AmpliSeq™ Library Kit 2.0 (Thermo Fisher Scientific, Massachusetts, USA) and then sequenced on Ion 318™ Chip Kit V2 using Ion PGM system (Thermo Fisher Scientific, Massachusetts, USA) for 500 cycles according to manufacturer's instructions. The raw signal data were analyzed using NextGENe Software Suite v3.4.2 (SoftGenetics, PA, USA) and in-house bioinformatics pipeline (BWA for alignment, GATK UnifiedGenotyper for calling and Annovar for annotation). The pipelines includes the alignment to the human genome 19 reference, some sequencing quality control (base quality score, mapping quality), coverage analysis and variant calling. After completion of the initial data analysis, lists of detected sequence variants (Single nucleotide variants [SNVs] and INDELs) were analyzed using variant filtering and annotation using COSMIC v.64 and dbSNP build 135. The variants were then filtered according to their frequency (>5% for SNVs and >10% for INDELs), reads coverage (>50× for SNVs and >100× for INDELs) and effect (nonsynonyme, splicing, not polymorphism).
Well characterized hot-spot mutations and gene amplification with ≥6 gene copy number were prevailed for treatment recommendation. Homozygous deletion or bi-allelic inactivation (inactivating mutation and heterozygous deletion) for tumor suppressor genes were taken into account. Passenger mutations or mutations known to be related to treatment resistance were taken into account only in respect of specific tumor types. KIT mutation already identified as therapeutic target in GIST were excluded from this analysis.
Genomic alterations were classified in signaling pathways according to the cancer genome atlas. Receptors of tyrosine kinase (RTK): KDR (VEGFR2), KIT, MET, PTPR, TIE1; Cell Cycle pathway: CCNE1 (cyclin E1), CDK4, CDKN2A (P16/INK4), RB1; PI3K pathway: PIK3CA, PTEN, RICTOR, TSC1, TSC2; RAS pathway: HRAS; p53 pathway: MDM2, TP53; Hedgehog pathway: PTCH1; Other non-classified genes: ADGRE2, BRCA2, DLEU2, DLEU7, DLG2, DMD, ELAVL2, HMGA2, LINGO2, MAP2K4, MAPK7, MYOCD, PMP22, TOP3A, TUSC7.
Molecular Tumor Board (MTB) and treatment decision
A weekly multidisciplinary Molecular Tumor Board (MTB) gathering together molecular biologists, medical oncologists, and pathologists reviewed NGS and aCGH reports in order to prioritize and select the genomic molecular alterations (amplifications, deletions, mutations) and allow recommendations for molecular-based therapies (i.e. actionable alterations). Decisions were based on an algorithm (Supplementary Table S2) defined using COSMIC database (Catalogue Of Somatic Mutation In Cancer) and the FATHMM (Functional Analysis Through Hidden Markov Models) score when variants of unknown functional and/or clinical significance were identified. Molecular-based recommended therapy (MBRT) was defined as a targeted therapy potentially active on the identified genomic alteration and recommended outside its EMA approval. Patients who initiated an MBRT were followed according to the clinical trial protocol they were referred to, or according to the routine medical practice for off label use of targeted therapies.
Statistical analysis
Categorical variables are expressed as percentages. Continuous variables are presented as median (range). OS was calculated as the time from the date of metastatic diagnosis to the date of death or last follow-up. Survival curves for OS were estimated using the Kaplan-Meier method. Median and 5-year survival rates were estimated. Univariate analyses used the Log-rank test for each variable of interest. A p-value of <0.05 was considered as statistically significant. PFS and OS for patients treated were calculated from the date of treatment initiation to the date of first progression or death for PFS and to the date of death or last follow up for OS. All statistical analyses were performed using R package version 3.5.2 (script available online). The results from the survival analyses are presented with the 95% confidence interval (CI).
Results
Patient and tumor characteristics
A total of 2579 locally advanced/metastatic cancer patients were included in the ProfiLER 01 study between February 2013 and February 2017 including 164 patients diagnosed with soft tissue sarcoma, bone sarcoma, or GIST. A molecular profile (i.e. at least one conclusive molecular analysis) was achieved for 158 patients. Molecular screening failures mostly occurred due to insufficient quality and/or quantity of tumor DNA with a failure rate of 4% (n = 6 from 164 patients included). Patient's characteristics at inclusion are detailed in Table 1. The median age at inclusion was 51.3 (6.8–84.6) years, whereas the median time between metastatic evolution and inclusion was 1.25 (0–17.7) years. A majority of women were included, 76% of patients had a performance status of 0 or 1. The median OS after metastatic diagnosis was 4.34 (95%CI 3.73–5.15) years, with a 5-year survival of 42% (95%CI 35–51%).
Table 1.
Patients characteristics.
| Median age at sarcoma diagnosis, years [IC 95%] | 48 [0–83] |
| Median age at locally advanced/metastatic diagnosis, years [IC 95%] | 50 [5–83] |
| Median age at inclusion, years [IC 95%] | 51.3 [6.8–84.6] |
| Sex: | |
| Female | 85 (58%) |
| Male | 73 (42%) |
| Performance status: | |
| 0 | 46 (29%) |
| 1 | 74 (47%) |
| 2 | 19 (12%) |
| 3 | 5 (3%) |
| NA | 14 (9%) |
| Tumor location: | |
| Trunk | 39 (25%) |
| Intra-abdominal | 68 (43%) |
| Limbs | 51 (32%) |
| Genomic profile: | |
| Single driver genomic alteration | 57 (36%) |
| Complex genomic profile | 101 (64%) |
| Median OS after metastatic diagnosis, years [IC 95%] | 4.34 [3.73–5.15] |
One hundred and one patients (64%) had a sarcoma with a complex genomic profile (leiomyosarcoma, osteosarcoma, chondrosarcoma, undifferentiated pleomorphic sarcoma, pleomorphic liposarcoma, embryonal rhabdomyosarcoma, unclassified sarcoma, MPNST, angiosarcoma, fibromyxoid sarcoma, hystiocytic sarcoma, follicular dendritic cells sarcoma, phyllode sarcoma) whereas 57 (36%) presented a sarcoma with a known single driver genomic abnormality (well differentiated liposarcoma, dedifferentiated liposarcoma, myxoid liposarcoma, endometrial stromal sarcoma, Ewing-sarcoma, synovial-sarcoma, GIST, alveolar rhabdomyosarcoma, epithelioid sarcoma, and dermatofibrosarcoma protuberans). The overall survival after the diagnosis of metastases was similar in these two groups: 4.23 (95%CI 3.23–5.4) years and 4.68 (3.38–6.58) years for sarcoma with complex genomic profile and sarcoma with single driver genomic abnormality, respectively.
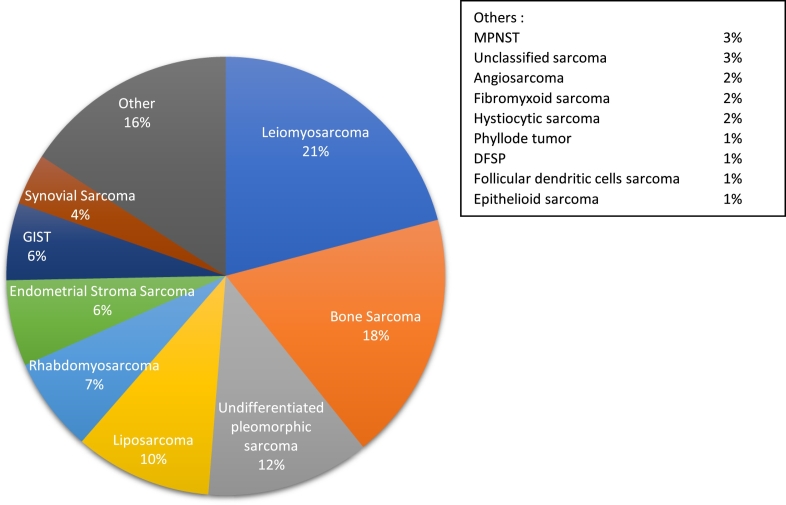
The intra-abdominal tumor location was the most frequent (43%). The mostly represented histological subtype was leiomyosarcoma (n = 33, 21%), 29 patients (18%) had a bone sarcoma (osteosarcoma, chondrosarcoma or Ewing-sarcoma), 19 patients (12%) an undifferentiated pleomorphic sarcoma (UPS), and 16 patients (10%) a liposarcoma. Histological subtype distribution is presented in Fig. 1.
Fig. 1.
Histological subtypes.
Genomic profiling was performed on archival tumor sample (n = 88) or on de novo biopsy for accessible lesion (n = 61), data were non-available in 9 samples.
Genomic alterations
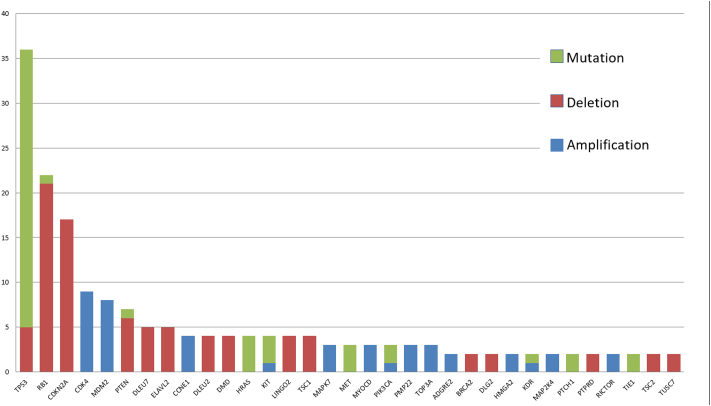
One hundred and six (67%) patients harbored at least one genomic alteration for a total of 289 genomic alterations in 143 different genes. Out of these 289 genomic alterations, 139 were homozygous deletions, 86 were gene amplifications, and 64 were somatic mutations. CHG array also identified gene breakpoint for 16 patients, suggesting an underlying rearrangement. In the 106 patients, sixty-eight (43%) presented multiple alterations, with up to 8 alterations for one patient. The median number of genomic alterations was 3.26 [1–8] and 2.26 [1–7] in patients with complex genomic and simple genomic sarcoma, respectively. Thirty-three out of 143 genes had more than one genomic alteration. The most frequently impacted genes were TP53, Rb1, CDKN2A, CDK4, MDM2, PTEN (Fig. 2). The incidence of molecular alterations in these 33 genes in each histological subtype are reported and classified according to the signaling pathway impacted (Table 2). The most frequently impacted genes were involved in cell cycle pathway especially in GIST, liposarcoma, and leiomyosarcoma, and genes involved in p53 pathway especially in liposarcoma and leiomyosarcoma.
Fig. 2.
Description of alterations for genes with more than one significant molecular alteration.
Table 2.
Incidence of actionable molecular alterations in each histological sub types and according to signaling pathway involved.
Only genes with more than one genomic alteration are presented in this table.
LMS = leiomyosarcoma, UPS = undifferentiated pleomorphic sarcoma, LPS = liposarcoma, RMS = rhabdomyosarcoma, ESS = endometrial stromal sarcoma, GIST = gastro-intestinal stroma sarcoma, SS = synovial sarcoma.
| LMS (n = 33) | Bone (n = 29) | UPS (n = 19) | LPS (n = 16) | RMS (n = 11) | ESS (n = 10) | GIST (n = 9) | SS (n = 6) | Others (n = 25) | % Total pop. | |
|---|---|---|---|---|---|---|---|---|---|---|
| RTK pathway | – | 10% | 5% | 6% | – | 10% | 33% | – | 16% | 8% |
| KDR (VEGFR2) | – | 3% | – | – | – | – | – | – | 4% | 1% |
| KIT | – | – | – | – | – | – | 22% | – | 8% | 3% |
| MET | – | 3% | – | 6% | – | – | – | – | 4% | 2% |
| PTPR | – | – | – | – | – | 10% | 11% | – | – | 1% |
| TIE1 | – | 3% | 5% | – | – | – | – | – | – | 1% |
| Cell cycle pathway | 42% | 21% | 37% | 50% | 9% | 30% | 67% | – | 28% | 33% |
| CCNE1 | – | 7% | 5% | 6% | – | – | – | – | – | 3% |
| CDK4 | – | 3% | – | 38% | 9% | 10% | – | – | – | 6% |
| CDKN2A (P16/INK4) | 3% | 7% | 5% | – | – | 10% | 67% | – | 24% | 11% |
| RB1 | 39% | 3% | 26% | 6% | – | 10% | – | – | 4% | 14% |
| PI3K pathway | 12% | 7% | 11% | 13% | 9% | 10% | 22% | – | 16% | 11% |
| PIK3CA | – | 3% | – | 6% | 9% | – | – | – | – | 2% |
| PTEN | 6% | – | 5% | 6% | – | – | – | – | 12% | 4% |
| RICTOR | 3% | 3% | – | – | – | – | – | – | – | 1% |
| TSC1 | 3% | – | – | – | – | – | 22% | – | 4% | 3% |
| TSC2 | – | – | 5% | – | – | 10% | – | – | – | 1% |
| RAS pathway | – | – | – | – | 9% | – | – | 33% | 4% | 3% |
| HRAS | – | – | – | – | 9% | – | – | 33% | 4% | 3% |
| p53 pathway | 42% | 17% | 32% | 75% | 9% | 20% | – | – | 16% | 28% |
| MDM2 | – | – | – | 44% | – | – | – | – | 4% | 5% |
| TP53 | 42% | 17% | 32% | 31% | 9% | 20% | – | – | 12% | 23% |
| Hedgehog pathway | – | – | 11% | – | – | – | – | – | – | 1% |
| PTCH1 | – | – | 11% | – | – | – | – | – | – | 1% |
| Other | 58% | 34% | 21% | 13% | – | 30% | 44% | 17% | 12% | 29% |
| ADGRE2 | – | 7% | – | – | – | – | – | – | – | 1% |
| BRCA2 | 3% | – | 5% | – | – | – | – | – | – | 1% |
| DLEU2 | 3% | – | 5% | – | – | 20% | – | – | – | 3% |
| DLEU7 | 9% | 3% | – | – | – | 10% | – | – | – | 3% |
| DLG2 | – | 3% | 5% | – | – | – | – | – | – | 1% |
| DMD | 3% | 3% | 5% | – | – | – | – | 17% | – | 3% |
| ELAVL2 | 3% | – | – | – | – | – | 22% | – | 8% | 3% |
| HMGA2 | – | – | – | 13% | – | – | – | – | – | 1% |
| LINGO2 | 3% | – | – | – | – | – | 22% | – | 4% | 3% |
| MAP2K4 | 3% | 3% | – | – | – | – | – | – | – | 1% |
| MAPK7 | 6% | 3% | – | – | – | – | – | – | – | 2% |
| MYOCD | 9% | – | – | – | – | – | – | – | – | 2% |
| PMP22 | 6% | 3% | – | – | – | – | – | – | – | 2% |
| TOP3A | 6% | 3% | – | – | – | – | – | – | – | 2% |
| TUSC7 | 3% | 3% | – | – | – | – | – | – | – | 1% |
The median overall survival after metastatic evolution was 4.51 (95%CI 3.86–6.02) years in patient without genomic actionable alteration, and 3.9 years (95%CI 3.16–5.07) in patient with at least one actionable genomic alteration, though no statistically significant difference was reached (p = 0.2).
Molecular based recommended therapy
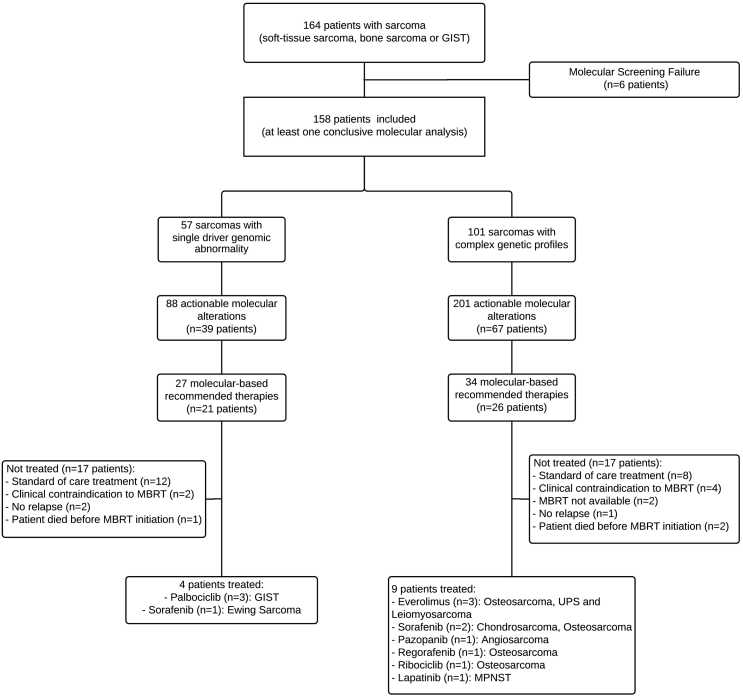
The MTB recommended MBRT for 47 patients (44% of the population with a genomic alteration, 30% of the total population), and a total of 61 MBRT were proposed. The most frequently recommended treatment were TKI (n = 23, 38%), then Cell Cycle inhibitors (n = 18, 29%), MTOR inhibitors (n = 12, 20%), MDMD2 inhibitors (n = 6, 10%), and PARP inhibitors (n = 2, 3%).
Thirteen patients (28% of the patients with MBRT, 8% of the total population) initiated the recommended treatment. For 34 patients, the MTB recommended a treatment that the patient didn't receive, in majority of cases because the use of a standard-of-care treatment was favored (n = 20), and for clinical contraindications (n = 6). The characteristics of the 13 patients treated are reported in Table 3. Nine patients had a complex genomic sarcoma and 4 had a sarcoma with a known single driver genomic abnormality (GIST [N = 3] and Ewing Sarcoma [N = 1]). Eight patients were oriented towards a molecularly tailored clinical trial based on the results of molecular screening. Five patients received anti-angiogenic tyrosine kinase inhibitor (sorafenib, pazopanib, regorafenib), four patients were treated with CDK4/6 inhibitors (ribociclib or palbociclib), 3 patients were treated with MTOR-inhibitors (everolimus), and 1 patient was treated with anti-Her1/2 tyrosine-kinase inhibitor (Fig. 3).
Table 3.
Characteristics of patients treated with MBRT.
| Age | Location | Histological subtype | Genomic alteration | MBRT initiated | Clinical trial | PFS (m) | Best response | OS (m) |
|---|---|---|---|---|---|---|---|---|
| 57 | Limbs | Leiomyosarcoma | AKT2 amplification | Everolimus | MOST | 2.6 | Progression | 10.9 |
| 47 | Trunk | MPNST | ERBB2 mutation c.G2329T/p.V777L | Lapatinib | MOST | 1.9 | Stable | 3.8 |
| 79 | Limbs | Angiosarcoma | FLT4 amplification | Pazopanib | MOST | 3.1 | Partial response | 10.7 |
| 30 | Limbs | UPS | AKT2 19q13.1-q13.2 deletion | Everolimus | – | 1.4 | Progression | 4.1 |
| 13 | Limbs | Osteosarcoma | KDR amplification | Regorafenib |
BAY15906 Phase I |
1.9 | Progression | 3.1 |
| 20 | Limbs | Osteosarcoma | VEGFA amplification | Sorafenib | – | 2.3 | Progression | 2.8 |
| 21 | Limbs | Osteosarcoma | PIK3CA amplification | Everolimus | MOST | 2.7 | Progression | 11 |
| 43 | Trunk | Osteosarcoma | CDK4 amplification | Ribociclib | LEE011 Phase I |
1.7 | Progression | 6.1 |
| 31 | Limbs | Ewing sarcoma | TIE1 mutation c.G1379C/p.S460T | Sorafenib | – | 3.6 | Stable | 7.4 |
| 58 | Limbs | Chondrosarcoma | KDR mutation c.3380C>A | Sorafenib | – | 33.6 | Stable | 39.6 |
| 58 | Abdominal | GIST | CDKN2A homozygous deletion | Palbociclib | – | 0.8 | Progression | 4.9 |
| 66 | Abdominal | GIST | CDKN2A homozygous deletion | Palbociclib | CycliGIST | 0.9 | Progression | 2.6 |
| 80 | Abdominal | GIST | CDKN2A homozygous deletion | Palbociclib | CycliGIST | 3.7 | Stable | 22.9 |
Fig. 3.
Flow diagram of patients included.
Patients receiving a targeted therapy based on genomic alteration had PFS of 2.3 (95%CI 1.7–not reached) months, and OS of 6.1 (95%CI 3.8–not reached) months. One objective response was observed: a patient with angiosarcoma reached partial response with pazopanib, though not lasting more than 3 months; four patients had a stable disease at first evaluation, including a long-lasting 33 months stabilization for one patient with chondrosarcoma treated with sorafenib. Eight patients had progressive disease. At data cut-off, all patients had stopped the recommended treatment for progression (Table 3).
Discussion
This study describes genomic alterations and molecular based recommended therapies in a cohort of 164 patients with locally advanced or metastatic sarcomas included in the ProfiLER 01 molecular screening program. Others teams reported comprehensive genomic analysis to identify relevant alterations in sarcomas [10,15]. However, ProfiLER 01 is the largest study to report potential efficacy of therapy recommended to target identified genomic alterations. The SHIVA and MOSCATO trials [17,18] included 8 patients with sarcoma and 26 patients with mesothelioma and sarcoma, respectively.
This analysis demonstrated that high throughput genomic analysis is feasible in sarcoma as in other tumors, with rare procedure-related failures, even for bone sarcoma known to be more challenging because of technical constraints resulting from poor quality samples (smaller size and degradation of nucleic acids due to the decalcification process).
The sarcoma cohort showed that 67% of patients had an identified molecular alteration, 30% of patients had molecular based recommended therapy and 8% initiated the proposed treatment, these proportions are similar to those reported in 2579 patients with various histological tumor types in the Profiler-01 study [19]. These results are also consistent with those of other genomics-guided therapy trials [10,12,17,18,20].
In these patients with sarcoma with identified alteration(s), the most frequent alterations occurred in TP53, in Rb1, and CDKN2A. Our series suggests that copy-number alterations may be more common than mutations in sarcomas, with the limitation of a sequencing panel covering mostly carcinoma-associated oncogenes and tumor suppressors and covering an overall limited number of DNA base pairs. Those findings are however consistent with those of TCGA [15] database, whereas NGS showed that mutations were the most frequent genomic alteration [10], and TP53 was the most frequent genomic alteration in a large cohort of sarcomas.
A MBRT could be offered for less than half of patients harboring an actionable molecular alteration (47 out of 106 patients). This low rate of MBRT proposal reflects the lack of access to innovative drugs, lack of molecular-based basket clinical trials aiming at assessing a potential predictive impact of molecular alteration for efficacy of targeted therapy. It also highlights the complexity of MTB decision-making, facing with multiple molecular alterations in a single tumor, variants of unknown functional and/or clinical significance or unknown predictive impacts of some hotspot variants (depending on tumor types).
Only 13 of the 47 patients for whom MBRT was proposed actually received the treatment. This low rate of treatment initiation may be partly explained by a molecular screening too lately proposed in advanced setting, usually after several lines of standard chemotherapy. We report a high proportion of patients with favorable prognosis at inclusion, patients in good condition for whom overall survival exceed 4 years. This cohort should not be considered as representative of “real life” patients with metastatic sarcoma. The ongoing study Profiler 02 aims at correcting this recruitment bias through molecular screening performed during the first line treatment to allow MBRT to be administrated in second line therapy.
In the thirteen (8% of the cohort) patients who initiated the MBRT, 4 patients had a simple genomic sarcoma and 9 a complex genomic sarcoma. Interestingly, 8 patients out of the 13 initiated MBRT within a clinical trial in which they could be included thanks to the molecular alteration identified. Among patients with sarcoma and single driver abnormalities, three patients with GIST harboring CDKN2A (p16INK4) homozygous deletion were treated with the CDK4/6 inhibitor palbociclib, and one patient with Ewing Sarcoma TIE1 mutated received sorafenib. Despite CDKN2A homozygous deletion, limited efficacy of CDK4/6 inhibitors has been recently reported in a phase 2 trial [21]. Among patients with sarcomas with complex genomic profile, bone sarcomas display several actionable alterations, mostly in RTK, cell cycle, and PI3K pathways. One patient with angiosarcoma harboring FLT4 amplification presented a good quality partial response to pazopanib. The patient was treated prior to drug approval. One patient with chondrosarcoma had long lasting stable disease during almost 3 years on sorafenib, as proposed MBRT to target an identified KDR mutation. These findings, limited by the low number of treated patients, suggest that molecular screening may be beneficial to complex genomics sarcoma, and would present marginal interest for sarcomas with single driver genomic abnormality.
Overall, our data suggest that molecular screening should not be used in routine practice but warrants further exploration in clinical trials. Similarly, the concept of personalizing cancer therapy needs to be further explored as newer drugs emerge, targeting different cellular processes (epigenetics).
Potential improvements in performance of such molecular screening program should nevertheless be considered. With more than half of genomic analyses performed on archived primary tumor samples, the frequency of genomic alterations may have been underestimated. Disease progression and metastatic evolution are known to correlate with the emergence of subclonal genomic alterations [22]. Moreover, biopsies may not adequately capture tumor heterogeneity. Whereas multiple biopsies are obviously restricted in clinical practice, reiterating a biopsy at disease progression or using liquid biopsies could strengthen genomic analysis results through enhanced representation of active tumoral subclones.
Some technical aspects could be considered to improve genomic screening yield. During the inclusion period of Profiler 01, the panel of genes has been enriched with ten additional genes in its second version (n = 69), but more genes could have been included for a more comprehensive analysis, such as ATRX [23], ARID1A or ARID1B [15], and INI1 [24]. The relevance of enlarged panel is raised in the ProfiLER 02 trial (NCT03163732), which randomizes the use of currently standard panel (87 genes) versus extended molecular profiling panel (324 genes). The use of larger panels may, in addition, allow the determination of tumor mutational burden to support the use of anti-PD1/PDL1.
In addition, molecular analyses in the ProfiLER 01 program were not initially designed to identify oncogenic fusions which have proved to be highly actionable molecular alterations, including in some subtypes of sarcomas [25]. Indeed fusions are not captured by conventional small scale NGS panels and require either a dedicated DNA NGS panel focusing on known breakpoint regions or RNA sequencing targeting oncogenes of interest. An increasing number of gene fusions are being discovered in sarcomas [26,27], and are mainly used for diagnostic purposes. The recent approval of the highly selective inhibitor of tropomyosin receptor kinases (TRK) larotrectinib in cancers harboring TRK fusion, shows that some of these fusions may be therapeutically relevant and may help improve patient outcome [28]. Contribution of whole transcriptome shotgun sequencing has not yet been demonstrated in routine use, but its impact on metastatic sarcoma patients' outcomes seems to be promising. However, large molecular analysis, using CGH array, NGS or RNA sequencing technologies are expensive and their cost-effectiveness have to be assessed attentively, with a need to limit healthcare costs and prioritize expenditures that improve patients outcomes.
The added value of large molecular screening for patients with single driver genomic abnormality is limited, whereas it may benefit to sarcomas with complex genomics, especially to bone sarcoma, enabling new treatments options.
Although we cannot recommend routine use of genomic screening, the use of wider panel and the addition of RNA sequencing and comprehensive genomic profiling in clinical trials will eventually allow a deeper understanding of sarcoma biology in humans and thus may impact treatment and patient outcome.
Funding
SIRIC-LYriCAN INCa-DGOS-Inserm_12563.
CRediT authorship contribution statement
Arnaud-Coffin Patrick: Conceptualization, Writing - Original Draft, Formal Analysis, Visualization Brahmi Mehdi: Writing - Review & Editing.
Vanacker Hélène: Writing - Review & Editing.
Eberst Lauriane: Writing - Review & Editing.
Tredan Olivier: Writing - Review & Editing, Supervision, Investigation.
Attignon Valery: Writing - Review & Editing, Data Curation.
Pissaloux Daniel: Data Curation, Formal Analysis.
Sohier Emilie: Data Curation, Formal Analysis.
Cassier Philippe: Writing - Review & Editing, Investigation.
Garin Gwenaelle: Data Curation, Formal Analysis.
Pérol David: Methodology, Formal Analysis.
Blay Jean-Yves: Writing - Review & Editing, Supervision.
Dufresne Armelle: Conceptualization, Writing - Original Draft, Visualization.
Declaration of competing interest
The authors have no conflict of interest to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100870.
Appendix A. Supplementary data
Supplementary material
References
- 1.WHO Classification of Tumours of Soft Tissue and Bone. Fourth Edition WHO - OMS. http://apps.who.int/bookorders/anglais/detart1.jsp?codlan=1&codcol=70&codcch=4005 (accessed March 2, 2019).
- 2.Stiller C.A., Trama A., Serraino D. Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. Eur. J. Cancer. 2013;49:684–695. doi: 10.1016/j.ejca.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Casali P.G., Abecassis N., Bauer S. Soft tissue and visceral sarcomas: ESMO–EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2018;29 doi: 10.1093/annonc/mdy096. (iv51–67) [DOI] [PubMed] [Google Scholar]
- 4.Casali P.G., Bielack S., Abecassis N. Bone sarcomas: ESMO–PaedCan–EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2018;29 doi: 10.1093/annonc/mdy310. (iv79–95) [DOI] [PubMed] [Google Scholar]
- 5.Casali P.G., Abecassis N., Bauer S. Gastrointestinal stromal tumours: ESMO–EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2018;29 doi: 10.1093/annonc/mdy320. (iv68–78) [DOI] [PubMed] [Google Scholar]
- 6.Vos M., Sleijfer S. EJC's biennial report on metastatic soft tissue sarcoma: state of the art and future perspectives. Eur. J. Cancer. 2018;88:87–91. doi: 10.1016/j.ejca.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Dufresne A., Brahmi M., Karanian M., Blay J.-Y. Using biology to guide the treatment of sarcomas and aggressive connective-tissue tumours. Nat. Rev. Clin. Oncol. 2018;15:443–458. doi: 10.1038/s41571-018-0012-4. [DOI] [PubMed] [Google Scholar]
- 8.Casali P.G., Zalcberg J., Le Cesne A. Ten-year progression-free and overall survival in patients with unresectable or metastatic GI stromal tumors: long-term analysis of the European Organisation for Research and Treatment of Cancer, Italian Sarcoma Group, and Australasian Gastrointestinal Trials Group intergroup phase III randomized trial on Imatinib at two dose levels. J. Clin. Oncol. 2017;35:1713–1720. doi: 10.1200/JCO.2016.71.0228. [DOI] [PubMed] [Google Scholar]
- 9.Rutkowski P., Van Glabbeke M., Rankin C.J. Imatinib mesylate in advanced dermatofibrosarcoma protuberans: pooled analysis of two phase II clinical trials. J. Clin. Oncol. 2010;28:1772–1779. doi: 10.1200/JCO.2009.25.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchesi C., Khalifa E., Laizet Y. Targetable alterations in adult patients with soft-tissue sarcomas: insights for personalized therapy. JAMA Oncol. 2018;4:1398–1404. doi: 10.1001/jamaoncol.2018.0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cote G.M., He J., Choy E. Next-generation sequencing for patients with sarcoma: a single center experience. Oncologist. 2018;23:234–242. doi: 10.1634/theoncologist.2017-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boddu S., Walko C.M., Bienasz S. Clinical utility of genomic profiling in the treatment of advanced sarcomas: a single-center experience. JCO Precis. Oncol. 2018:1–8. doi: 10.1200/PO.18.00096. [DOI] [PubMed] [Google Scholar]
- 13.Groisberg R., Hong D.S., Holla V. Clinical genomic profiling to identify actionable alterations for investigational therapies in patients with diverse sarcomas. Oncotarget. 2017;8 doi: 10.18632/oncotarget.16845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris M.H., DuBois S.G., Glade Bender J.L. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: the Individualized Cancer Therapy (iCat) study. JAMA Oncol. 2016;2:608. doi: 10.1001/jamaoncol.2015.5689. [DOI] [PubMed] [Google Scholar]
- 15.Abeshouse A., Adebamowo C., Adebamowo S.N. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–965.e28. doi: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquart J., Chen E.Y., Prasad V. Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA Oncol. 2018;4:1093. doi: 10.1001/jamaoncol.2018.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massard C., Michiels S., Ferté C. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7:586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 18.Tourneau C.L., Delord J.-P., Gonçalves A. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 19.Trédan O., Wang Q., Pissaloux D. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann. Oncol. 2019;30:757–765. doi: 10.1093/annonc/mdz080. [DOI] [PubMed] [Google Scholar]
- 20.Gounder M.M., Ali S.M., Robinson V. Impact of next-generation sequencing (NGS) on diagnostic and therapeutic options in soft-tissue and bone sarcoma. J. Clin. Oncol. 2017;35:11001. [Google Scholar]
- 21.Toulmonde M., Blay J.-Y., Bouche O. Activity and safety of palbociclib in patients with advanced gastrointestinal stromal tumors refractory to imatinib and sunitinib: a biomarker-driven phase II study. Clin. Cancer Res. 2019;25:4611–4615. doi: 10.1158/1078-0432.CCR-18-3127. [DOI] [PubMed] [Google Scholar]
- 22.Jamal-Hanjani M., Wilson G.A., McGranahan N. Tracking the evolution of non–small-cell lung cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 23.Michaud M., Pérot G., Lesluyes T. The emerging role of ATRX and chromatin remodeling in pleomorphic sarcomas oncogenesis. Eur. J. Cancer. 2016;61:S28. [Google Scholar]
- 24.Kohashi K., Oda Y. Oncogenic roles of SMARCB1/INI1 and its deficient tumors. Cancer Sci. 2017;108:547–552. doi: 10.1111/cas.13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 26.Hofvander J., Tayebwa J., Nilsson J. RNA sequencing of sarcomas with simple karyotypes: identification and enrichment of fusion transcripts. Lab. Investig. 2015;95:603–609. doi: 10.1038/labinvest.2015.50. [DOI] [PubMed] [Google Scholar]
- 27.Mertens F., Johansson B., Fioretos T., Mitelman F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 28.Drilon A., Laetsch T.W., Kummar S. Efficacy of larotrectinib in fusion–positive cancers in adults and children. N. Engl. J. Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material