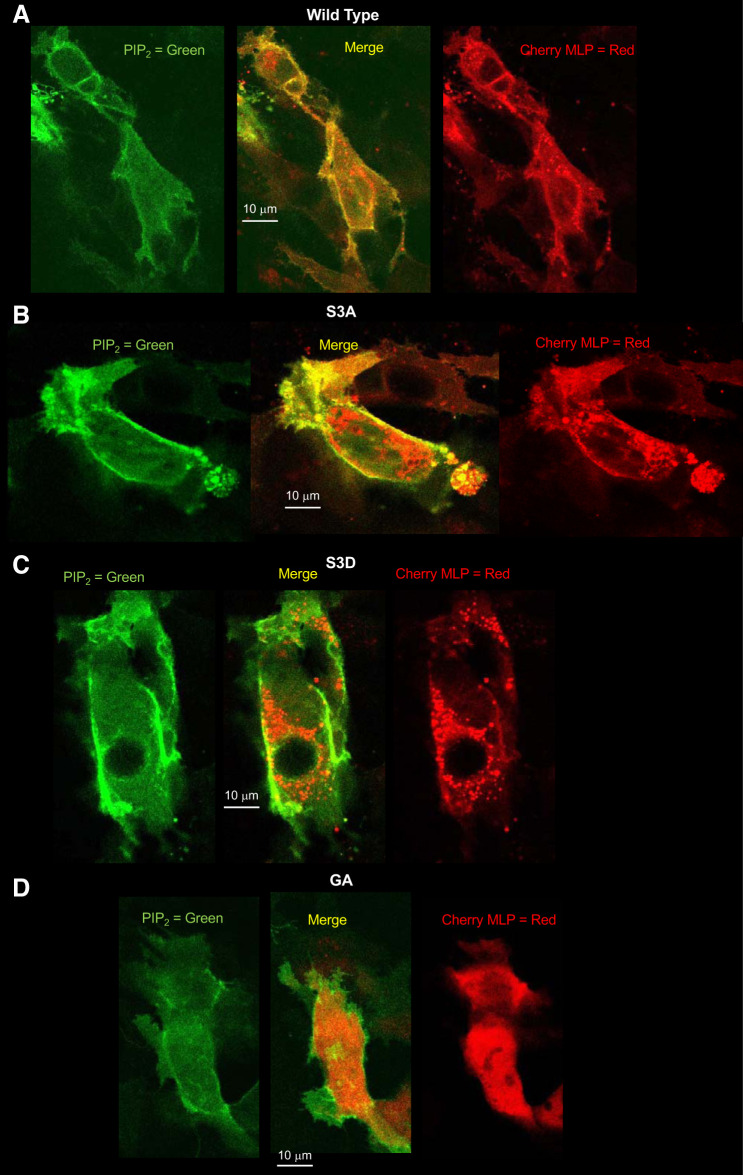
Fig. 9.
A–D: differential cellular distribution of myristoylated alanine-rich protein kinase C substrate-like protein-1 (MLP-1) mutants. To examine the cellular localization of the different MLP-1 constructs, we transfected DCT-15 cells (a distal convoluted tubule clonal cell line) with a phosphatidylinositol 4,5-bisphosphate (PIP2) reporter and each of the MLP-1 constructs and then used multiphoton microscopy to produce z-axis stacks of the transfected cells. A: an examination of the merged image of the wild-type construct shows that MLP-1 strongly colocalizes with PIP2 (yellow pixels at the membrane); however, quantitative analysis with ImageJ (Coloc 2 module) shows that only 25% of pixels in the image colocalize. Examination of green pixels shows that 64% colocalize with red pixels, but 62% of red pixels do not colocalize with green pixels. This is consistent with the prediction that most green pixels (PIP2) are at the membrane and many of them are associated with red pixels (wild-type MLP-1), but there are many red pixels in the cytosol. B: for S3A, 58% of all pixels colocalize. Ninety-nine percent of green pixels are associated with red pixels, and 96% of red pixels are associated with green pixels. This is consistent with the S3A mutant being predominantly associated with the membrane with little in the cytosol compared with wild-type MLP-1. C: for S3D, only 13% of red and green pixels are colocalized with 71% of green pixels unassociated with red pixels. This reflects the distribution of most of S3D in the cytosol with only a small fraction associated with the membrane. D: the GA construct has 22% green and red colocalized pixels. This is close to the colocalization of the wild-type construct, implying that the myristoylation does not strongly influence MLP-1 association with the membrane. Images are typical of 3 similar experiments.