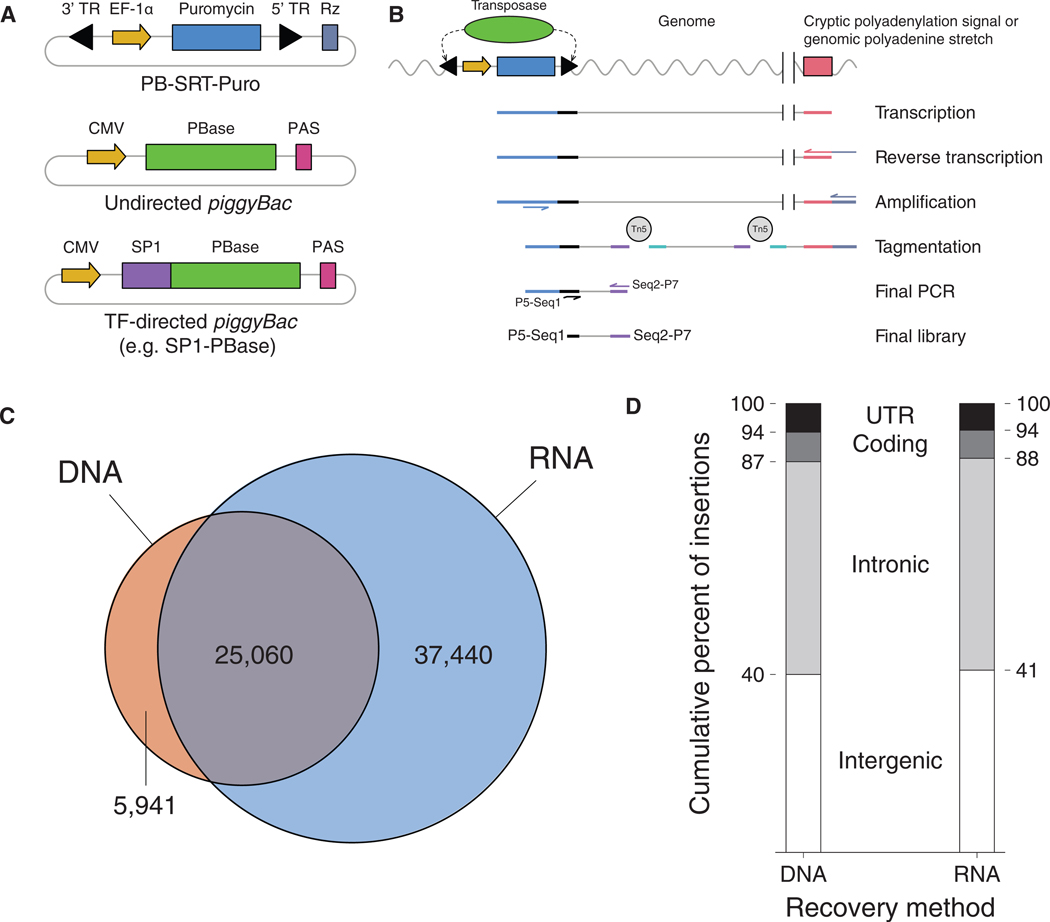
Figure 1. Self-Reporting Transposons Are Mapped More Efficiently from RNA Compared to DNA.
(A) Schematics of a self-reporting piggyBac transposon with puromycin reporter gene (PB-SRT-Puro) and undirected (PBase) and SPI-directed (SP1-PBase) piggyBac transposases.
(B) Molecular workflow for mapping SRTs from bulk RNA libraries.
(C) Overlap of SRTs recovered by DNA- or RNA-based protocols in HCT-116 cells.
(D) Distribution of insertions with respect to genetic annotation between SRT libraries prepared from either DNA or RNA. TR, terminal repeat; Puro, puromycin; PAS, polyadenylation signal.