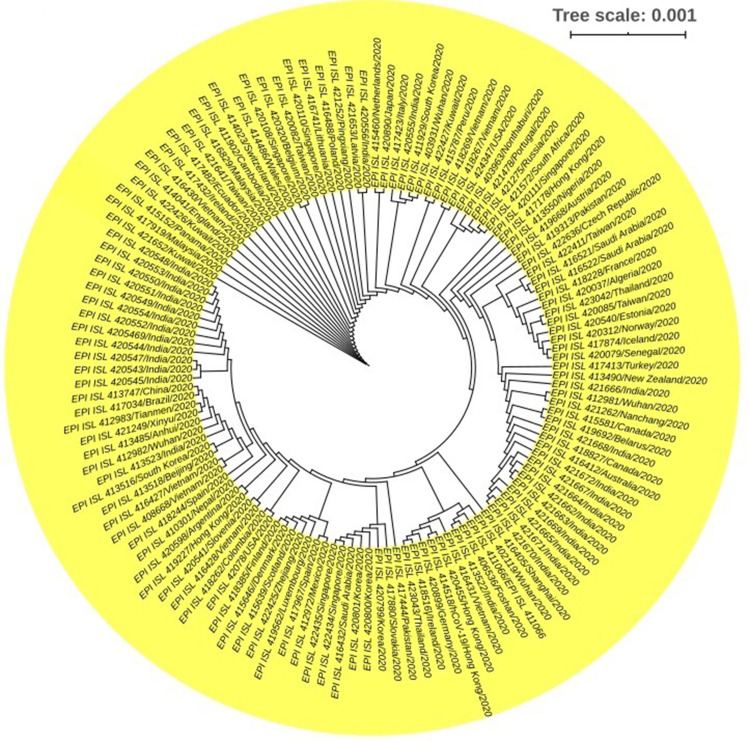
Figure 2. Phylogenetic tree of SARS-CoV-2.
200 complete genome sequences of SARS-CoV-2 retrieved from global initiative on sharing all influenza data (GISAID) (https://www.gisaid.org/) from different countries were used to build this tree. The sequences were aligned using MAFFT online server (Katoh, Rozewicki & Yamada, 2019), and a maximum likelihood tree was built with iTOL (interactive Tree Of Life). Each node represents a single strain which is found to be patient and/or sample specific, and not clustered according to geographical locations. Tree scale 0.01, represents days before the time of lastly sampled genomes by scale*365.