Abstract
N6-methyladenosine (m6A) is regarded as the most abundant, prevalent and conserved internal mRNA modification in mammalian cells. M6A can be catalyzed by m6A methyltransferases METTL3, METTL14 and WTAP (writers), reverted by demethylases ALKBH5 and FTO (erasers), and recognized by m6A -binding proteins such as YTHDF1/2/3, IGF2BP1/2/3 and HNRNPA2B1 (readers). Emerging evidence suggests that m6A modification is significant for regulating many biological and cellular processes and participates in the pathological development of various diseases, including tumors. This article reviews recent studies on the biological function of m6A modification and the methylation modification of m6A in urological tumors.
Keywords: N6-methyladenosine (m6A), writers, erasers, readers, urological tumors
Introduction
In past decades, epigenetic modification has been identified to be involved in diverse biological processes and disease progression, attracting more and more attention. Epigenetics is a study of reversible, inheritable phenotypes that do not involve changes in nuclear DNA sequences (Mohammad et al., 2019), and primarily includes RNA interference, histone modification, chromatin rearrangement, DNA methylation and RNA modification (Arguello et al., 2019; McGee and Hargreaves, 2019).
RNA modification was previously regarded as occurring in high-abundance RNA species, while emerging evidence indicates that it is characterized in lowly abundant species of RNA such as non-coding RNAs and Mrna (Dominissini, 2014; Li X. et al., 2016). Among them, RNA methylation has attracted accumulating attention in recent years and N6-methyladenosine (m6A) is the most prevalent RNA methylation sites (Pan, 2013). M6A modification was firstly reported to be interrelated to the regulation of gene expression, growth and development in 1970s (Desrosiers et al., 1974; Perry et al., 1975; Chandola et al., 2015; Hsu et al., 2017), and it has been regarded as one of the most common mRNA modifications recently. Many researches have revealed that m6A modification mainly occured in the consensus sequence RRACH sequence (R = A, G; H = A, C, U) (Li L. J. et al., 2018), which is enriched in stop codons, 3′ untranslated region (UTR) and the last exon in non-coding RNA (Dominissini et al., 2012; Meyer et al., 2012). Besides, m6A is widespread in RNA of bacteria, viruses and eukaryotes (Desrosiers et al., 1974; Wang Y. et al., 2014; Deng et al., 2015; Fu et al., 2015; Greer et al., 2015; Zhang et al., 2015; Liu J. et al., 2016; Zhu et al., 2018).
M6A modification is reversible and catalyzed by many relevant enzymes (Batista, 2017; Dai et al., 2018). Studies have shown that m6A is involved in various biological and disease processes via regulating target gene expression (Chen X.Y. et al., 2019; Lan et al., 2019). M6A modification is associated with various diseases, such as neurological diseases (Liu E. Y et al., 2017; Salta and De Strooper, 2017) and cancers. In this review, we provide a broad overview of the relationship between RNA m6A methylation and urological tumors. We further highlight the possible uses in diagnostic, prognostic and therapeutic applications of m6A modifications for urological tumors.
Regulators of m6A
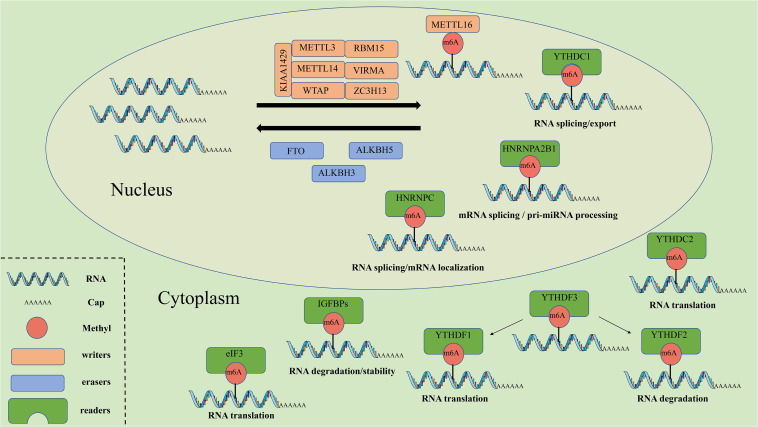
Similar to histone modification and DNA methylation, m6A modification is reversible and dynamic, and influences biological functions that are primarily mediated by three types of regulators: methyltransferases (“writers”), demethylases (“erasers”) and m6A binding proteins (“readers”). The methyltransferase complex (MTC) can catalyze m6A, demethylase can remove m6A, while RNA reader proteins can recognize m6A and bind to the RNA. These proteins play an essential biological role in m6A modifications (Table 1, Figure 1). Cross-talk among writers, erasers and readers of m6A is involved in the development and progression of tumors (Deng et al., 2018; Panneerdoss et al., 2018).
TABLE 1.
Functions of m6A regulators in RNA metabolism.
| Type | m6A Regulators | Function | References |
| m6A writer | METTL3 | Catalyzes m6A modification | Schwartz et al., 2014 |
| Zhou J. et al., 2015 | |||
| – | METTL14 | Forms a stable complex with METTL3 | Schwartz et al., 2014 |
| – | – | – | Zhou J. et al., 2015 |
| METTL16 | Catalyzes m6A modification | Warda et al., 2017 | |
| – | WTAP | Contributes to the localization of METTL3-METTL14 heterodimer to the nuclear speckle | Ping et al., 2014 |
| RBM15 | Binds the m6A complex and recruit it to special RNA site | Moindrot et al., 2015 | |
| – | VIRMA | Recruits the m6A complex to the special RNA site and interacts with polyadenylation | Wang T. et al., 2020 |
| – | – | Cleavage factors CPSF5 and CPSF6 | – |
| – | – | – | – |
| ZC3H13 | Bridges WTAP to the mRNA-binding factor Nito | Wen et al., 2018 | |
| m6A eraser | FTO | Mediates demethylation of both hm6A and f6A in mRNA | Basak et al., 2019 |
| – | – | – | – |
| ALKBH5 | Removes m6A modification | Tang et al., 2018 | |
| m6A reader | YTHDF1 | Facilitates mRNA translation efficiency | Liu J. et al., 2020 |
| – | – | – | – |
| YTHDF2 | Promotes mRNA degradation | Zhou J. et al., 2015 | |
| – | YTHDF3 | Enhances translation and degradation by interacting with YTHDF1 and YTHDF2 | Shi et al., 2017 |
| – | – | – | Li A. et al., 2017 |
| YTHDC1 | Recruits the RNA splicing and controls the nuclear export | Roundtree et al., 2017b | |
| – | YTHDC2 | Interacts with RNA helicase and increases the translation efficiency of target RNA | Mao et al., 2019 |
| – | – | – | – |
| IGF2BPs | Recruits RNA stabilizers | Huang H. et al., 2018 | |
| – | HNRNPA2B1 | Mediates mRNA splicing and primary microRNA processing | Alarcon et al., 2015 |
| – | – | – | – |
| HNRNPC | Influences alternative splicing and mRNA localization | Guichard et al., 2012 | |
| – | EIF3 | Facilitates cap-independent translation | Meyer et al., 2015 |
FIGURE 1.
The molecular mechanism of m6A. M6A can be installed by “writers” (METTL3/14/16, WTAP, RBM15, VIRMA, KIAA1429, and ZC3H13), removed by “erasers” (FTO, ALKBH5, and ALKBH3), and recognized by “readers” (YTHDF1/2/3, YTHDC1/2, IGF2BPs, HNRNPs, and eIF3). METTL3/14/16, methyltransferase like 3/14/16; WTAP, WT1 associated protein; RBM15, RNA binding motif protein 15; VIRMA, vir like m6A methyltransferase associated; ZC3H13, zinc finger CCCH-type containing 13; FTO, FTO alpha-ketoglutarate dependent dioxygenase; ALKBH5, alkB homolog 5, RNA demethylase; ALKBH3, alkB homolog 3, RNA demethylase; YTHDF1/2/3, YTH N6-methyladenosine RNA binding protein 1/2/3; YTHDC1/2, YTH domain containing 1/2; IGF2BPs, insulin like growth factor 2 mRNA binding proteins; HNRNPs, heterogeneous nuclear ribonucleo proteins; eIF3, eukaryotic translation initiation factor 3 subunit.
Methyltransferases (“Writers”)
MTC has been identified to regulate the installation of m6A and Methyltransferase-like 3 (METTL3), METTL14, and Wilms tumor 1-associated protein (WTAP) have been proved as the core components of this complex (Ping et al., 2014; Schwartz et al., 2014; Zhou J. et al., 2015). METTL3 is an Sadenosyl methionine (SAM)-binding protein and regarded as a major catalytic enzyme with functions reminiscent of the N6-adenine methyltransferase system (Barbieri et al., 2017). Besides, METTL3 is highly conserved in eukaryotes from yeast to humans (Bokar et al., 1997). WTAP can also increase the binding ability of METTL3, thus regulating recruitment of the complex to mRNA targets (Ping et al., 2014). METTL14 could form a stable complex with METTL3 and both of them contain a SAM-binding motif. With the help of WTAP, METTL3-METTL14 could colocalize in nuclear speckles and form a heterodimer, so as to participate in catalytic activity (Liu J. et al., 2014; Zhao X. et al., 2014). Besides, VIRMA, RBM15, ZC3H13 and KIAA1429 are the new components of the m6A “writer” complex (Moindrot et al., 2015; Wang X. et al., 2016; Deng et al., 2018; Wen et al., 2018).
METTL3
The writer METTL3 has been identified to be involved in various biological processes. METTL3 can enhance the BAT-mediated adaptive thermogenesis and suppress obesity and systemic insulin resistance via targeting the 3′ UTR of the PRDM16, PPARG, and UCP1 transcript to install the m6A modification (Wang Y. et al., 2020). The ablation of METTL3 in germ cells severely inhibited spermatogonial differentiation and blocked the initiation of meiosis (Xu et al., 2017). Besides, METTL3 was also shown to be upregulated in various solid tumors and associated with poor prognosis. In oral squamous cell carcinoma (OSCC), METTL3 can facilitate tumor growth and metastasis through making an increment in m6A modification and expression of c-Myc transcript (Zhao W. et al., 2020). In colorectal cancer (CRC), METTL3 stabilizes HK2 and GLUT1 expression via a m6A -IGF2BP2/3-dependent mechanism (Shen et al., 2020). Additionally, METTL3 might affect tumor metastasis through promoting the maturation of pri-miR-1246 (Peng et al., 2019). METTL3 enhances the splicing of precursor miR-143-3p and facilitates its biogenesis, thereby promoting the brain metastasis of lung cancer (LC) (Wang H. et al., 2019). Moreover, METTL3 induces non-small cell lung cancer (NSCLC) drug resistance and metastasis by promoting Yes-associated protein (YAP) mRNA translation via a m6A -YTHDF1/3/eIF3b-dependent mechanism (Jin D. et al., 2019). In gastric cancer (GC), overexpression of METTL3 can promote the stability of ZMYM1, thereby enhancing epithelial mesenchymal transformation (EMT) process and tumor metastasis (Yue et al., 2019). In addition, upregulated METTL3 facilitates GC growth and liver metastasis through installing m6A modifications of HDGF transcript (Wang Q. et al., 2020).
METTL14
Studies have demonstrated that METTL14 is associated with a lower risk for development of neoplasms. In CRC, METTL14 acts as a tumor-suppressor to inhibit cell growth and metastasis in vitro and in vivo. Mechanistical study demonstrated that downregulated METTL14 substantially abolishes m6A modifications of XIST and augments XIST expression (Yang X. et al., 2020). In addition, METTL14 can inhibit CRC cell proliferation, migration and invasion via the miR-375-YAP1/SP1 signal axis (Chen X. et al., 2020). Although both of METTL3 and METTL14 could act as m6A “writer”, METTL3 might promote the progression of CRC, while METTL14 functions as a tumor suppressor in CRC. METTL14 can also assume an oncogenic role in triple-negative breast cancer (TNBC) (Shi et al., 2020), pancreatic cancer (Kong et al., 2020) and leukemia (Weng et al., 2018). Moreover, METTL14 is significantly upregulated in Epstein–Barr virus (EBV) latently infected cells. METTL14 can lead to oncogenesis via increasing m6A modifications of the indispensable EBV latent antigen EBNA3C and thus facilitating its stability and expression. Interestingly, EBNA3C can also enhance stability and expression of METTL14 (Lang et al., 2019).
METTL16
METTL16 has been recently shown to have distinct target RNAs for m6A modification. Studies have revealed that METTL16 can bind a subset of mRNAs and methylate U6 small nuclear RNA (U6 snRNA) and long non-coding RNA (lncRNA) (Brown et al., 2016; Fitzsimmons and Batista, 2019). Moreover, the UACAGAGAA sequence is essential for METTL16-mediated-methylation and the Nterminal module of METTL16 is required for RNA binding (Doxtader et al., 2018; Mendel et al., 2018). METTL16 is involved in catalyzing m6A in A43 of the U6 small nuclear RNA (Warda et al., 2017). Under loss-of-SAM conditions, METTL16 can induce the splicing of a retained intron, thereby enhancing level of MAT2A and expression of SAM, while down-regulation of METTL16 and YTHDC1 can abolish SAM-responsive regulation of MAT2A (Pendleton et al., 2017; Shima et al., 2017). While the specific role of METTL16 in solid tumors remain to be further explored.
Demethylases (“Erasers”)
The reversible and dynamic m6A modification can be mediated by obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) (m6A “erasers”) (Jia et al., 2011; Zheng et al., 2013). Both FTO and ALKBH5 are members of the ALKB family of dioxygenases. As the first reported demethylase, FTO can also mediate demethylation of both N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A) in mRNA (Basak et al., 2019). ALKBH5 plays an essential role in mRNA export and RNA metabolism (Tang et al., 2018).
FTO
As an m6A eraser, FTO is associated with the initiation and development of various cancers including hepatocellular carcinoma (HCC), melanoma, breast cancer and glioma. In HCC, SIRT1 destabilizes FTO and thus steering the m6A of downstream elements and consecutive mRNA expression in tumorigenesis (Liu X. et al., 2020). In melanoma, FTO can impair IFNγ-induced killing via augmenting CXCR4, PD-1 and SOX10 expression via repressing YTHDF2-mediated degradation and suppress response to anti-PD-1 blockade immunotherapy (Yang S. et al., 2019). In breast cancer, FTO enhances breast cancer cell growth, colony formation and metastasis. Mechanistical study demonstrated that FTO can mediate m6A demethylation of BNIP3 transcript and induce its degradation via an YTHDF2 independent mechanism (Niu et al., 2019). The ethyl ester form of meclofenamic acid (MA2) inhibits FTO and enhances the effect of the chemotherapy drug temozolomide (TMZ) on suppressing proliferation of glioma cells (Xiao et al., 2020).
ALKBH5
ALKBH5 has been regarded as a tumor suppressor in many cancers. In NSCLC, ALKBH5 suppresses cell growth and metastasis both in vitro and in vivo via repressing miR-107/LATS2-mediated YAP activity and YTHDFs-mediated YAP expression (Jin D. et al., 2020). In pancreatic cancer, downregulated ALKBH5 predicts poor prognosis and knockdown of ALKBH5 markedly facilitates tumor growth and metastasis (Tang et al., 2020). In HCC, ALKBH5 is characterized as a tumor suppressor and could attenuate the expression of LYPD1 via an mA-dependent manner in HCC cells (Chen Y. et al., 2020). In addition, ALKBH5 can augment steady-state CYR61 mRNA expression via an m6A -dependent mechanism, thereby repressing trophoblast invasion (Li X. C. et al., 2019).
m6A Binding Proteins (“Readers”)
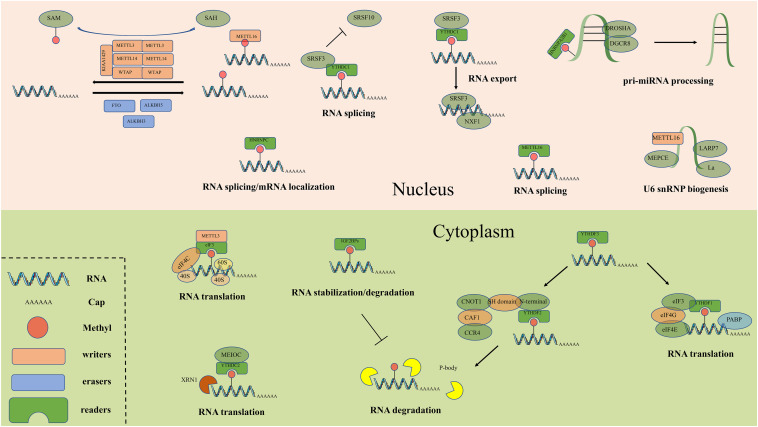
M6A readers can recognize and bind to m6A sites and regulate target RNA translation, splicing, nuclear export and decay (Figure 2). In YTH (YT521-B homology) domain family, the evolutionarily conserved YTH domain acts as the module for directly binding to m6A. YTHDF1–3 and YTHDC1–2 are the main five YTH domain proteins. YTHDF1 can bind to m6A sites around the stop codon and thus facilitating mRNA translation efficiency (Liu X. et al., 2020). YTHDF2 can accelerate degradation and deadenylation of the transcripts by bringing m6A-modified translatable mRNAs to mRNA decay sites and recruiting CCR4-NOT deadenylase complex (Zhou J. et al., 2015). YTHDF3 can, respectively, promote RNA translation through associating with YTHDF1 and enhance RNA degradation by interacting with YTHDF2 (Li A. et al., 2017; Shi et al., 2017). In contrast to the prevailing model, where each DF paralog binds to distinct subsets of mRNAs, Zaccara and Jaffrey show that the DF paralogs bind proportionately to each m6A site throughout the transcriptome (Zaccara and Jaffrey, 2020). YTHDC1 recruits the RNA splicing and control the nuclear export (Roundtree et al., 2017b). YTHDC2 interacts with RNA helicase and increases the translation efficiency of target RNA (Mao et al., 2019). The insulin-like growth factor 2 mRNA binding protein (IGF2BP) family proteins, including IGF2BP1-3, can recognize m6A containing transcripts. IGF2BPs exert their functions via recruiting RNA stabilizers (Huang H. et al., 2018). Eukaryotic initiation factor 3 (EIF3) can facilitate cap-independent translation (Meyer et al., 2015). Heterogeneous nuclear ribo nucleo protein (HNRNP) family proteins include hnRNPC, hnRNPG and hnRNPA2B1. HnRNPC and hnRNPG can influence alternative splicing and mRNA localization (Guichard et al., 2012) while hnRNPA2B1 can bind to m6A -containing primary microRNAs and enhance microRNA maturation (Alarcon et al., 2015).
FIGURE 2.
The detailed molecular mechanism of m6A enzymes. The “writers”, “erasers” and “readers” relay on a variety of related factors install, remove and recognize m6A mutation and participate in RNA metabolic processes, including translation, splicing, export, degradation and so on.
YTHDF1
More recently, YTHDF1 has been proved to be upregulated in various tumors, associated with more advanced stages and poorer survival. In ovarian cancer, YTHDF1 promotes tumor growth and metastasis. Mechanistically, YTHDF1 binds to the m6A modification site of EIF3C 3′-UTR to increase the translation of EIF3C mRNA (Liu X. et al., 2020). YTHDF1 could promote the translation of frizzled7 (FZD7) in an m6A-dependent manner, leading to hyper-activation of the Wnt/β-catenin pathway and promotion of gastric carcinogenesis (Pi et al., 2020). Besides, YTHDF1 binds the m6A modification site of Robo3.1 3′-UTR and promotes its translation in an m6A-independent mechanism. While down-expression of YTHDF1 in spinal commissural neurons contributes to pre-crossing axon guidance defects (Zhuang M. et al., 2019).
YTHDF2
Evidence has shown that YTHDF2 can act as an oncogene or tumor suppressor in different tumor models. In HCC, YTHDF2 decreased expression level is associated with poor prognosis and classification. YTHDF2 may participate in the occurrence and progression of HCC by processing the decay of m6A-containing serpin family E member 2 (SERPINE2) and interleukin 11 (IL11) mRNAs (Hou et al., 2019). Besides, YTHDF2 can suppress tumor growth through modulating the m6A methylation of EGFR mRNA by the m6A/mRNA degradation pathway. However, YTHDF2 promotes the cancer stem cell liver phenotype and cancer metastasis by binding m6A-modified OCT4 mRNA (Zhang et al., 2020). YTHDF2 can also interact with miRNA, miR-145 targets YTHDF2 and results in its degradation (Yang Z. et al., 2017). Moreover, YTHDF2 is also involved in the initation of other biological process. In spermatogenesis, YTHDF2 regulates cell proliferation and adhesion via modulating the m6A methylation of MMPs and simultaneously decreasing the overall translational output (Huang T. et al., 2020). Knockdown of YTHDF2 promotes the expression of MAP2K4 and MAP4K4 and activates MAPK and NF-κB signaling pathways, which facilitate the expression of proinflammatory cytokines and exacerbate the inflammatory response in LPS-stimulated RAW 264.7 cells (Yu et al., 2019).
YTHDF3
YTHDF3 has been reported to play a fine-tuning role in the RNA accessibility of YTHDF1 and YTHDF2 and biological process. In CRC, lncRNA GAS5 leads to ubiquitin-mediated degradation of YAP via interacting with WW domain of YAP, thus repressing tumor progression. While YTHDF3 might recognize m6A-modified GAS5 and induce decay of it (Ni et al., 2019). YTHDF3 can serve as a negative regulator to enhance the translation of FOXO3 mRNA, thereby maintaining host antiviral immune function and preventing inflammatory response (Zhang Y. et al., 2019).
YTHDC1
YTHDC1 and YTHDC2 have conserved m6A binding domain and preferentially bind to m6A-modified RNA in RRm6ACH consensus sequence (Roundtree et al., 2017a). YTHDC1 is involved in processing of pre-mRNA transcripts of F6, SRSF3, and SRSF7 in the oocyte nucleus, and it may play a crucial role in fetal development (Kasowitz et al., 2018). MAT2A mRNA can be methylated by METTL16 and YTHDC1 can bind to the m6A modification site of MAT2A 3′-UTR. Downregulation of METTL16 and YTHDC1 might effectively abolish SAM-responsive regulation of MAT2A (Shima et al., 2017). The m6A modification site of long non-coding RNA X-inactive specific transcript (XIST) can be preferentially read by YTHDC1 and it’s required for XIST function (Patil et al., 2016). Recent study shows that the ability of the YTH domain of YTHDC1 binding to ssDNA is stronger than in an RNA context. However, the YTH domains of YTHDF2 and YTHDF1 exhibit the opposite effect (Woodcock et al., 2020).
YTHDC2
YTHDC2 could bind mitotic transcripts, specific piRNA precursors and interact with RNA granule components, licensing the proper progression of germ cells through meiosis (Bailey et al., 2017). YTHDC2 results in colon cancer metastasis through augmenting translation of HIF-1α, it may be a potential diagnostic marker and therapy target in colon cancer (Tanabe et al., 2016). YTHDC2 binds to the mRNA of lipogenic genes and participates in the regulation of hepatic lipogenesis and TG homeostasis (Zhou B. et al., 2020).
IGF2BPs
IGFBPs could use common RNA binding domains to recognize m6A containing transcripts and play a significant role in many diseases. In breast cancer, FGF13-AS1 can reduce the half-life of c-Myc (Myc) mRNA by binding IGF2BPs, thus suppressing cell proliferation, migration and invasion (Ma et al., 2019). In ovarian cancer, IGF2BP1 enhances cell proliferative and invasive ability by antagonizing miRNA-impaired gene expression, the elevate expression of IGF2BP1 is correlated to poor prognosis (Muller et al., 2018). IGF2BP1 could function as an adaptor protein to recruit the CCR4-NOT complex, so as to initiate the degradation of the lncRNA highly up-regulated in liver cancer (HULC) (Hammerle et al., 2013). In pancreatic cancer, IGF2BP2 could promote cell growth through activating the PI3K/Akt signaling pathway and be negatively regulated by miR-141 (Xu et al., 2019). In addition, IGF2BP2 enhances cancer stemness-like properties and promotes tumorigenesis by acting as a reader for m6A modified DANCR (Hu et al., 2020). In gastric cancer, miR-34a directly targets IGF2BP3, overexpression of IGF2BP3 promotes cell proliferation and invasion (Zhou Y. et al., 2017). IGF2BP3 could interact with RNA-binding protein Lin28b and thereby promotes stability and expression of target mRNAs such as B-cell regulators Pax5 and Arid3a, so as to participate in the fetal–adult hematopoietic switch (Wang S. et al., 2019).
EIF3
EIF3 is crucial for specialized translation initiation via interacting with the 5′; cap region, resulting in assemblage of translation initiation complexes on eIF3-specialized mRNA (Lee et al., 2016). Study has proved that YTHDF1 might promote the translation of EIF3 via recognizing the m6A-modified sites of EIF3 mRNA and simultaneously augments the overall translational output, thus facilitating tumorigenesis and metastasis in ovarian cancer (Liu X. et al., 2020). In renal cell carcinoma (RCC), knockdown of EIF3 dramatically decreases cell viability with sunitinib treatment. Mechanistically, EIF3 could interact with GRP78 and enhance protein stability by blocking the ubiquitin-mediated degradation of GRP78 (Huang H. et al., 2019). In gallbladder cancer (GBC), EIF3 can stabilize GRK2 protein through blocking ubiquitin-mediated degradation, wherefore activating PI3K/Akt signal pathway and enhancing tumor growth and metastasis (Zhang et al., 2017). All above studies demonstrate that EIF3 is a vital role in the progerssion of various cancers.
Roles of RNA m6A in Urological Tumors
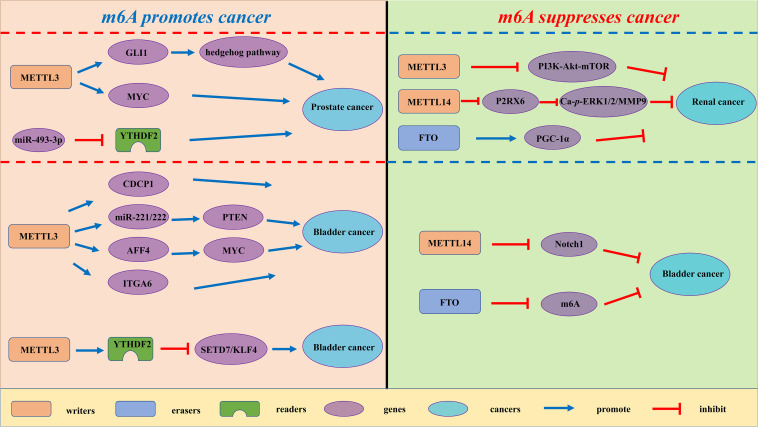
Accumulating evidence indicates that RNA m6A modification is related to the tumorigenesis, development and progression of urological tumors. Therefore, we summarize these latest advances of m6A modification in urological tumors (Table 2, Figure 3).
TABLE 2.
The roles of RNA m6A in urological tumors.
| Cancer | m6A Regulators | Role in cancer | Biological function | Mechanism | References |
| Renal cancer | METTL3 | Suppressor | Suppresses RCC proliferation, migration | Regulates EMT and PI3K-Akt-mTOR pathways | Rini et al., 2009 |
| gene | and invasion | ||||
| – | METTL14 | Suppressor | Suppresses RCC migration and invasion | Down-regulates P2RX6 protein translation | Li X. et al., 2017 |
| – | – | gene | – | – | – |
| – | – | – | – | – | – |
| FTO | Suppressor | Spppress RCC growth | Promotes PGC-1α expression by reducing m6A levels | Gong et al., 2019 | |
| gene | |||||
| Prostate cancer | METTL3 | Oncogene | Promotes PCa growth and metastasis | Regulates hedgehog pathway | Siegel et al., 2020 |
| – | – | – | – | – | – |
| METTL3 | Oncogene | Promotes PCa proliferation, migration | Promotes MYC expression by increasing m6A levels | Cai et al., 2019 | |
| and invasion | |||||
| – | YTHDF2 | Oncogene | Promotes PCa proliferation and migration | / | Yuan et al., 2020 |
| – | – | – | – | – | – |
| Bladder cancer | METTL3 | Oncogene | Promotes BC growth | Promotes CDCP1 mRNA modification and translation | Bray et al., 2018 |
| – | METTL3 | Oncogene | Promotes BC proliferation | Interactes with the microprocessor protein DGCR8 and | Yang F. et al., 2019 |
| – | – | – | – | positively modulates the pri-miR221/222 process | – |
| – | – | – | – | – | – |
| METTL3 | Oncogene | Promotes BC growth and metastasis | Regulates AFF4/NF-κB/MYC signaling network | Han et al., 2019 | |
| – | METTL3 | Oncogene | Promotes BC growth and metastasis | promote the translation of ITGA6 mRNA | Cheng et al., 2019 |
| – | – | – | – | – | – |
| METTL3/YTHDF2 | Oncogene | Promotes BC growth and metastasis | METTL3/YTHDF2 may mediate the mRNA decay | Jin H. et al., 2019 | |
| of tumor suppressors SETD7 and KLF4 | |||||
| – | METTL14 | Suppressor | Promotes the proliferation, self-renewal, metastasis | METTL14 knockdown may enhance the RNA | Xie et al., 2020 |
| – | – | gene | and tumor initiating capacity of bladder TICs | stability of Notch1 mRNA | – |
| – | – | – | – | – | – |
| FTO | Suppressor | Inhibits BC proliferation and migration | / | Gu et al., 2019 | |
| Gene |
FIGURE 3.
The potential roles of m6A in urological tumors progression. The potential role of m6A in urological tumors progression is reflected in the regulation of tumor-associated gene expression. M6A modification promotes urological tumors progression by enhancing oncogene expression and inhibiting tumor suppressor gene expression. M6A modification hinders cancer progression by inhibiting oncogene expression and enhancing tumor suppressor gene expression.
Renal Cell Carcinoma
Renal cell carcinoma (RCC) is derived from renal epithelium and is one of the most common cancers worldwide, making up nearly 2% to 3% of all adult malignancies (Rini et al., 2009). Li and collaborators demonstrated that METTL3 was a potential prognostic marker of RCC, and the expression levels of METTL3 are interrelated to tumor size and histological grade. Inhibition of METTL3 could obviously promote cell proliferation, migration and invasion, and make cell cycle arrest (Li X. et al., 2017). In addition, METTL3 knockdown might activate oncogenic PI3K/Akt/mTOR signaling pathway. Hence, METTL3 might function as a tumor suppressor in the tumorigenesis of RCC. Gong and co-workers found that the expression level of METTL14 is decreased in RCC (Gong et al., 2019). Additionally, the mRNA level of METTL14 is associated with RCC patients’ overall survival. Knockdown of METTL14 promotes the mRNA and protein expression levels of P2RX6, while P2RX6 could further regulate the Ca2+-mediated p-ERK1/2/MMP9 signal pathway promote cell migration and invasion. Zhuang C. et al. (2019) found that PGC-1α underwent m6A methylation in RCC. As an m6A demethylase, FTO could recognize the m6A sites of PGC-1α and reduce its methylation level, therefore leading to the increases in mitochondria biogenesis and oxidative phosphorylation and the decreases in tumor growth of RCC (Zhuang C. et al., 2019).
Prostate Cancer
Prostate cancer (PCa) has been regarded as the most common cancer among men and the second cancer-related deaths in the men in 2019 (Siegel et al., 2020). Despite recent advances in many therapies, the 5 years’ survival rate for prostate cancer patients remains low. Cai et al. found that METTL3 is overexpressed in PCa tissues and cell lines (Cai et al., 2019). Elevated expression of METTL3 could promote cell proliferation, survival, colony formation, and invasion. Moreover, knockdown of METTL3 could decrease the m6A modification and expression of GLI1, thereby regulating hedgehog pathway. Yuan et al. (2020) demonstrated that the mRNA expression level of METTL3 was increased in prostate cancer tissues. Additionally, the expression level of METTL3 is associated with the deterioration of PCa patients’ condition. Mechanistically, METTL3 could enhance MYC (c-myc) expression via elevating m6A levels of MYC mRNA transcript, so as to facilitate the proliferative, migrative and invasive ability of cancer cells. Li found that YTHDF2, an m6A reader, was upregulated in prostate cancer tissues and cell lines (Li J. et al., 2018). Knockdown of YTHDF2 led to decreased levels of m6A and impaired proliferation and migration of PCa cells. Therefore, YTHDF2 played a vital role in the initition and progression of PCa.
Bladder Cancer
Bladder cancer (BCa) is the most common urogenital and the 10th most common cancer worldwide, with an estimated 549 000 new cases and 200 000 deaths in 2018 (Bray et al., 2018). Despite the improvement of clinical diagnosis and therapies, BCa is regarded as a major cause of cancer-interrelated morbidity and mortality. In the study of Yang F. et al. (2019) the expression levels of METTL3 were elevated in BCa patient samples. The increase in METTL3 expression was proven to be correlated with BCa growth and progression in vitro and in vivo. Moreover, METTL3 could positively regulate CDCP1 process based on an m6A -dependent mode, bringing about elevated expression of CDCP1. Han et al. (2019) demonstrated that the expression level of METTL3 in BCa was significantly up-regulated and associated with poor prognosis of BCa patients. They found that METTL3 might interact with the microprocessor protein DGCR8 and positively modulate the pri-miR221/222 process through an m6A -dependent mechanism. Cheng and coworkers elucidated that METTL3 was obviously up-regulated in BCa tissues and significantly promoted growth and metastasis of BCa (Cheng et al., 2019). Mechanistically, METTL3 might promote BCa progression via AFF4/NF-κB/MYC signaling pathway. Jin and coworkers demonstrated that METTL3 and ALKBH5 can alter cell adhesion via regulating ITGA6 expression in BCa (Jin H. et al., 2019). Increased m6A methylation enhanced the translation of ITGA6 mRNA by binding of YTHDF1 and YTHDF3 and promoted malignant phenotypes in BCa. Xie and coworkers found that knockout of METTL3 impaired tumor growth and metastasis, METTL3/YTHDF2 m6A axis could directly degrad the mRNA expression of the tumor suppressors SETD7 and KLF4, leading to the development and progression of BCa (Xie et al., 2020). Gu et al. (2019) found a decrease of N6-methyladenosine in BCa and bladder tumor initiating cells (TICs). In addition, METTL14 is down-regulated in BCa and bladder TICs and it could promote the proliferation, metastasis, self-renewal and enhance tumor initiating capacity of bladder TICs. Mechanistically, METTL14 might regulate Notch1 expression in an m6A-dependent manner. Wen demonstrated that knockdown of FTO could accelerate the progression of BCa (Wen et al., 2020), while the potential mechanism remains unknown.
Testicular Germ Cell Tumors
Testicular germ cell tumors (TGCTs) are the most common solid neoplasm among men aged between 14 and 44 years (Cheng et al., 2018). Despite the advanced prognosis of localized TGCTs, approximately 20–30% of patients may experience disease recurrence during surveillance (Mortensen et al., 2016). Lobo and coworkers demonstrated that abundance of m6A and expression of VIRMA/YTHDF3 were different among TGCTs subtypes, with higher levels in seminomas (SEs), suggesting a contribution to SE phenotype maintenance (Lobo et al., 2019). However, the potential biological roles of VIRMA/YTHDF3 remain to be further explored.
Wilms Tumor
Wilms tumor (WT) is the most prevalent childhood kidney tumor characterized by the disorganized and dysregulated development of a kidney (Davidoff, 2009; Servaes et al., 2019). Hua et al. (2020) found an obvious relationship between ALKBH5 rs1378602 AG/AA genotypes and decreased Wilms tumor risk in children in clinical stage I diseases. However, the observed association should be further validated in another well-designed analysis with other larger ethnicities.
Potential Application of RNA M6A in Urological Tumors Rna
RNA m6A as Biomarker in Urological Tumors
Mounting evidence has indicated that m6A regulators have the potential to be superior diagnostic and prognostic biomarkers for urological tumors patients. Strick et al. conducted qRT-PCR to detect the gene expressions of ALKBH5 and FTO were studied in 166 ccRCC and 106 normal renal tissues. They found that the expression level of ALKBH5 and FTO were obviously decreased in ccRCC tissues (Strick et al., 2020). Declined mRNA levels of ALKBH5 and FTO were related to a shortened overall and cancer-specific survival following nephrectomy. Therefore, ALKBH5 and FTO could be used as prognostic biomarkers for RCC. Zhao Y. et al. (2020) demonstrated that METTL14 mRNA expression negatively correlated with the RCC stages and positively correlated with RCC patients’ overall survival, it might be a potential biomarker of RCC. Yuan et al. performed the qRT-PCR to detect the mRNA expression level of METTL3 in 84 clinical human PCa specimens and 32 corresponding adjacent normal specimens. The results showed that a significant positive association between METTL3 expression was observed with tumor stage and metastasis. Moreover, the expression level of METTL3 had remarkable prognostic value for overall survival and disease-free survival (Yuan et al., 2020); hence, METTL3 might play a vital role in PCa progression and metastasis. Chen et al. concluded that m6A regulators were related to malignant clinicopathological features of BCa and a risk signature with FTO, WTAP and YTHDC3 might play vital roles in diagnosis and prognosis of BCa patients (Chen M. et al., 2019). In TGCTs, VIRMA and YTHDF3 might be prognostic factors (Lobo et al., 2019).
RNA m6A as Therapeutic Targets in Urological Tumors
The critical roles of m6A in urological tumors suggest that it has the potential to be involved in tumor therapy. A number of studies have indicated that m6A modification is significant in therapies of urological tumors, especially in targeted treatment. Zhuang et al. found that the Von Hippel-Lindau (VHL) -deficient cells expressing FTO might restore mitochondrial activity, induce oxidative stress and ROS production and suppressed tumor growth, via promoting PGC-1α expression by decreasing m6A levels in its mRNA transcripts (Zhuang C. et al., 2019). Therefore, the m6A methylation and m6A-related regulators, and uncovers an essential FTO-PGC-1α axis might play a vital role in the treatment of RCC. Gong and coworkers found that ATP could enhance cell migration and invasion via rugulating P2RX6 expression in RCC (Gong et al., 2019). Mechanistically, ATP-P2RX6 could modulate the Ca2+-mediated p-ERK1/2/MMP9 signal pathway, while METTL14 might down-regulate P2RX6 protein translation in an m6A-dependent manner. Herein, the further exploration of regulation of METTL14 expression might contribute to develop a new approach to repress RCC progression. Li et al. suggested that METTL3 expression is higher in PCa than in normal prostate tissues, especially in PCa with bone metastasis (Li E. et al., 2020). METTL3 regulates the expression of Integrin β1 (ITGB1) through m6A-HuR-dependent mechanism, which affects the binding of ITGB1 to Collagen I and tumor cell motility, so as to promote the bone metastasis of PCa. Therefore, METTL3 might act as a therapeutic target for PCa bone metastasis. Wen et al. found that knockdown of FTO could enhance cell proliferation and migration and protect BCa cells from cisplatin-induced cytotoxicity (Wen et al., 2020). Hence, targeting the m6A modification of FTO may be beneficial to the treatment of BCa.
Discussion
Recently, RNA epigenetics is emerging as a hot topic. Among them, m6A modification has become a new layer of post-transcriptional regulation of gene expression. The implications of m6A modifications in human carcinogenesis have been verified in many kinds of cancers, including urological tumors. In this review, we summarized the potential biological effects of m6A-related regulators, and particularly focused on the impacts of m6A modification on different tumors in the urinary system. M6A can be installed by the methyltransferase, while these modifications may be removed by m6A eraser demethylases. Furthermore, m6A readers could specifically recognize the m6A methylation sites and thus regulating mRNA splicing, translation, degradation, nuclear export, and other cellular processes. Besides, m6A methylation and its related regulatory factors are reported to be involved in the processing and the biological function of non-coding coding RNAs (Coker et al., 2019; Huang H. et al., 2020).
However, m6A methylation seems to serve as a double-edged sword due to the specific mechanism for m6A in cancers remains unknown. Some genes may lead to cancer progression after m6A methylation, while removal of m6A modification can result in the progression of other tumors. For example, in HCC, sumo1 modification of METTL3 can promote tumor progression via regulating snail mRNA homeostasis (Xu et al., 2020), while in glioblastoma, LncRNA SOX2OT can facilitate temozolomide resistance through promoting SOX2 expression via ALKBH5-mediated epigenetic regulation (Liu X. et al., 2020). In addition, the same m6A-associated regulator may play crucial roles in the same type of cancer via targeting different downstream genes. For instance, in CRC, METTL3 can promote tumor progression through enhancing the expression of either MYC (Xiang et al., 2020) or CCNE1 (Zhu W. et al., 2020). Additionally, researches have reported conflicting findings in the same type of cancer; for instance, in CRC, METTL3 and METTL14 play totally opposite roles in tumor initition and progression (Li T. et al., 2019; Yang X. et al., 2020). Overall, all above studies show that m6A methylation and its related regulatory networks are complex and need to be further explored. Moreover, Han et al. found that METTL3 can enhance tumor growth of BCa through accelerating pri-miR221/222 maturation based on m6A-dependent mode (Han et al., 2019), while Gu et al. (2019) reported that METTL14 can inhibit bladder tumorigenesis through N6-methyladenosine of Notch1. The above discrepancy may result from several factors such as case sample size and different related regulatory genes. Furthermore, studies have identified the therapeutic potential of m6A modification. METTL3 might induce NSCLC drug resistance and metastasis via modulating the MALAT1-miR-1914-3p-YAP axis (Jin D. et al., 2019). In glioma, METTL3 can promote glioma radioresistance and stem-like cell maintenance (Visvanathan et al., 2018). In melanoma, FTO can act as an m6A demethylase to promote melanoma tumorigenesis and anti-PD-1 resistance (Yang S. et al., 2019). R-2HG can inhibit FTO activity and thus elevating m6A mRNA modification in R-2HG-sensitive leukemia cells, thereby generating anti-leukemia effects (Su et al., 2018). In cervical squamous cell carcinoma (CSCC), FTO can regulate the chemo-radiotherapy resistance by targeting β-catenin through mRNA demethylation (Zhou S. et al., 2018).
The advanced development of m6A modification study marks a novel insight in the dignosis and therapy of various diseases. Nevertheless, we believe that future prospects on m6A modification need to be further explored. Firstly, several databases (such as GEPIA, TCGA et al.) were used in many studies to explore the prognostic significance of m6A regulators expression in OS and DFS of urological tumors patients. Hence, expansion of the sample size and screening factors are essential for early diagnosis and prognosis; while the specificity and sensitivity of m6A-related regulators also need to be discussed. Secondly, more and more clinical pratice are urgent for confirming the therapeutic potential of m6A regulatory factors and related pathways. Thirdly, it’s significant to construct a complex and specific regulatory network model of m6A and its associated modifiers in a single cancer. Fourthly, exploring other components of m6A methylation and demethylation and effectors is necessary.
Conclusion
Urological tumors are major public health concern with growing prevalence. Studies have showed that m6A methylation plays a significant role in prevention, treatment and management of various urological tumors; however, more endeavors and more multi-center and large-scale research are urgent for exploring the relationship between m6A modification and urological tumors.
Author Contributions
YL and YG collected the related manuscript and finished the manuscript and figures. RJ gave constructive guidance and made final approval. LX, ZX, and QD participated in the design of this review. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from the National Natural Science Foundation of China (NSFC) (Grant Numbers: 81570613, 81370853, and 81802531), Jiangsu Provincial Social Development Project (BE2017615), and Jiangsu Provincial Medical Innovation Team (2016).
References
- Alarcon C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello A. E., Leach R. W., Kleiner R. E. (2019). In vitro selection with a site-specifically modified RNA library reveals the binding preferences of N(6)-methyladenosine reader proteins. Biochemistry 58 3386–3395. 10.1021/acs.biochem.9b00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A. S., Batista P. J., Gold R. S., Chen Y. G., de Rooij D. G., Chang H. Y., et al. (2017). The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6:e26116. 10.7554/eLife.26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millan-Zambrano G., Robson S. C., et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552 126–131. 10.1038/nature24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak E. A., Koolen S. L. W., Hurkmans D. P., Schreurs M. W. J., Bins S., Oomen-de Hoop E., et al. (2019). Correlation between nivolumab exposure and treatment outcomes in non-small-cell lung cancer. Eur. J. Cancer 109 12–20. 10.1016/j.ejca.2018.12.008 [DOI] [PubMed] [Google Scholar]
- Batista P. J. (2017). The RNA modification N(6)-methyladenosine and its implications in human disease. Genomics Proteomics Bioinformatics 15 154–163. 10.1016/j.gpb.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J. A., Shambaugh M. E., Polayes D., Matera A. G., Rottman F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Brown J. A., Kinzig C. G., DeGregorio S. J., Steitz J. A. (2016). Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 113 14013–14018. 10.1073/pnas.1614759113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Yang F., Zhan H., Situ J., Li W., Mao Y., et al. (2019). RNA m(6)A Methyltransferase METTL3 promotes the growth of prostate cancer by regulating hedgehog pathway. Onco Targets Ther. 12 9143–9152. 10.2147/OTT.S226796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandola U., Das R., Panda B. (2015). Role of the N6-methyladenosine RNA mark in gene regulation and its implications on development and disease. Brief. Funct. Genomics 14 169–179. 10.1093/bfgp/elu039 [DOI] [PubMed] [Google Scholar]
- Chen M., Nie Z. Y., Wen X. H., Gao Y. H., Cao H., Zhang S. F. (2019). m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci. Rep. 39:BSR20192892. 10.1042/BSR20192892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu M., Xu X., Zeng K., Liu X., Sun L., et al. (2020). METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol. Ther. 28 599–612. 10.1016/j.ymthe.2019.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen X. Y., Zhang J., Zhu J. S. (2019). The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18:103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhao Y., Chen J., Peng C., Zhang Y., Tong R., et al. (2020). ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol. Cancer 19:123. 10.1186/s12943-020-01239-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Albers P., Berney D. M., Feldman D. R., Daugaard G., Gilligan T., et al. (2018). Testicular cancer. Nat. Rev. Dis. Primers 4:29. 10.1038/s41572-018-0029-0 [DOI] [PubMed] [Google Scholar]
- Cheng M., Sheng L., Gao Q., Xiong Q., Zhang H., Wu M., et al. (2019). The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene 38 3667–3680. 10.1038/s41388-019-0683-z [DOI] [PubMed] [Google Scholar]
- Coker H., Wei G., Brockdorff N. (2019). m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 1862 310–318. 10.1016/j.bbagrm.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Dai D., Wang H., Zhu L., Jin H., Wang X. (2018). N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 9:124. 10.1038/s41419-017-0129-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff A. M. (2009). Wilms’ tumor. Curr. Opin. Pediatr. 21 357–364. 10.1097/MOP.0b013e32832b323a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Chen K., Luo G. Z., Weng X., Ji Q., Zhou T., et al. (2015). Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 43 6557–6567. 10.1093/nar/gkv596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Su R., Weng H., Huang H., Li Z., Chen J. (2018). RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 28 507–517. 10.1038/s41422-018-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., Rottman F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71 3971–3975. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D. (2014). Genomics and proteomics. Roadmap to the epitranscriptome. Science 346:1192. 10.1126/science.aaa1807 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Doxtader K. A., Wang P., Scarborough A. M., Seo D., Conrad N. K., Nam Y. (2018). Structural basis for regulation of METTL16, an S-Adenosylmethionine homeostasis factor. Mol. Cell 71 1001–1011.e4. 10.1016/j.molcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons C. M., Batista P. J. (2019). It’s complicated. m(6)A-dependent regulation of gene expression in cancer. Biochim. Biophys. Acta Gene Regul. Mech. 1862 382–393. 10.1016/j.bbagrm.2018.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Luo G. Z., Chen K., Deng X., Yu M., Han D., et al. (2015). N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161 879–892. 10.1016/j.cell.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D., Zhang J., Chen Y., Xu Y., Ma J., Hu G., et al. (2019). The m(6)A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca(2+) influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J. Exp. Clin. Cancer Res. 38:233. 10.1186/s13046-019-1223-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Blanco M. A., Gu L., Sendinc E., Liu J., Aristizabal-Corrales D., et al. (2015). DNA Methylation on N6-Adenine in C. elegans. Cell 161 868–878. 10.1016/j.cell.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Wang Z., Zhou N., Li G., Kou Y., Luo Y., et al. (2019). Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol. Cancer 18:168. 10.1186/s12943-019-1084-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I. B., et al. (2012). Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44 694–698. 10.1038/ng.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle M., Gutschner T., Uckelmann H., Ozgur S., Fiskin E., Gross M., et al. (2013). Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 58 1703–1712. 10.1002/hep.26537 [DOI] [PubMed] [Google Scholar]
- Han J., Wang J. Z., Yang X., Yu H., Zhou R., Lu H. C., et al. (2019). METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18:110. 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Zhang H., Liu J., Zhao Z., Wang J., Lu Z., et al. (2019). YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 18:163. 10.1186/s12943-019-1082-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. J., Shi H., He C. (2017). Epitranscriptomic influences on development and disease. Genome Biol. 18:197. 10.1186/s13059-017-1336-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Peng W. X., Zhou H., Jiang J., Zhou X., Huang D., et al. (2020). IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 27 1782–1794. 10.1038/s41418-019-0461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua R. X., Liu J., Fu W., Zhu J., Zhang J., Cheng J., et al. (2020). ALKBH5 gene polymorphisms and Wilms tumor risk in Chinese children: a five-center case-control study. J. Clin. Lab. Anal. 34:e23251. 10.1002/jcla.23251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Gao Y., Liu A., Yang X., Huang F., Xu L., et al. (2019). EIF3D promotes sunitinib resistance of renal cell carcinoma by interacting with GRP78 and inhibiting its degradation. EBioMedicine 49 189–201. 10.1016/j.ebiom.2019.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Weng H., Chen J. (2020). m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell 37 270–288. 10.1016/j.ccell.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Liu Z., Zheng Y., Feng T., Gao Q., Zeng W. (2020). YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m(6)A/mRNA pathway. Cell Death Dis. 11:37. 10.1038/s41419-020-2235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D., Guo J., Wu Y., Du J., Yang L., Wang X., et al. (2019). m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 12:135. 10.1186/s13045-019-0830-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jin D., Guo J., Wu Y., Yang L., Wang X., Du J., et al. (2020). m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol. Cancer 19:40. 10.1186/s12943-020-01161-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Ying X., Que B., Wang X., Chao Y., Zhang H., et al. (2019). N(6)-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine 47 195–207. 10.1016/j.ebiom.2019.07.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowitz S. D., Ma J., Anderson S. J., Leu N. A., Xu Y., Gregory B. D., et al. (2018). Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14:e1007412. 10.1371/journal.pgen.1007412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F., Liu X., Zhou Y., Hou X., He J., Li Q., et al. (2020). Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int. J. Biochem. Cell Biol. 122:105731. 10.1016/j.biocel.2020.105731 [DOI] [PubMed] [Google Scholar]
- Lan Q., Liu P. Y., Haase J., Bell J. L., Huttelmaier S., Liu T. (2019). The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 79 1285–1292. 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- Lang F., Singh R. K., Pei Y., Zhang S., Sun K., Robertson E. S. (2019). EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 15:e1007796. 10.1371/journal.ppat.1007796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. S., Kranzusch P. J., Doudna J. A., Cate J. H. (2016). eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536 96–99. 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Chen Y. S., Ping X. L., Yang X., Xiao W., Yang Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27 444–447. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Wei B., Wang X., Kang R. (2020). METTL3 enhances cell adhesion through stabilizing integrin beta1 mRNA via an m6A-HuR-dependent mechanism in prostatic carcinoma. Am. J. Cancer Res. 10 1012–1025. [PMC free article] [PubMed] [Google Scholar]
- Li J., Meng S., Xu M., Wang S., He L., Xu X., et al. (2018). Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget 9 3752–3764. 10.18632/oncotarget.23365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. J., Fan Y. G., Leng R. X., Pan H. F., Ye D. Q. (2018). Potential link between m(6)A modification and systemic lupus erythematosus. Mol. Immunol. 93 55–63. 10.1016/j.molimm.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Li T., Hu P. S., Zuo Z., Lin J. F., Li X., Wu Q. N., et al. (2019). METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer 18:112. 10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Tang J., Huang W., Wang F., Li P., Qin C., et al. (2017). The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget 8 96103–96116. 10.18632/oncotarget.21726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xiong X., Yi C. (2016). Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14 23–31. 10.1038/nmeth.4110 [DOI] [PubMed] [Google Scholar]
- Li X. C., Jin F., Wang B. Y., Yin X. J., Hong W., Tian F. J. (2019). The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics 9 3853–3865. 10.7150/thno.31868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zhou J., Wang C., Chi Y., Wei Q., Fu Z., et al. (2020). LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 11:384. 10.1038/s41419-020-2540-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Cali C. P., Lee E. B. (2017). RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 10 509–518. 10.1242/dmm.028613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li K., Cai J., Zhang M., Zhang X., Xiong X., et al. (2020). Landscape and regulation of m(6)A and m(6)Am Methylome across human and mouse tissues. Mol. Cell 77 426–440.e6. 10.1016/j.molcel.2019.09.032 [DOI] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhu Y., Luo G. Z., Wang X., Yue Y., Wang X., et al. (2016). Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 7:13052. 10.1038/ncomms13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wei Q., Jin J., Luo Q., Liu Y., Yang Y., et al. (2020). The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 48 3816–3831. 10.1093/nar/gkaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Liu J., Xiao W., Zeng Q., Bo H., Zhu Y., et al. (2020). SIRT1 regulates N(6) -methyladenosine RNA modification in hepatocarcinogenesis by inducing RANBP2-dependent FTO SUMOylation. Hepatology. 10.1002/hep.31222 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Lobo J., Costa A. L., Cantante M., Guimaraes R., Lopes P., Antunes L., et al. (2019). m(6)A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: a role in seminoma phenotype maintenance. J. Transl. Med. 17:79. 10.1186/s12967-019-1837-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Liu X., Zhou S., Li W., Liu C., Chadwick M., et al. (2019). Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 450 63–75. 10.1016/j.canlet.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Mao Y., Dong L., Liu X. M., Guo J., Ma H., Shen B., et al. (2019). m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10:5332. 10.1038/s41467-019-13317-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee S. L., Hargreaves M. (2019). Epigenetics and exercise. Trends Endocrinol. Metab. 30 636–645. 10.1016/j.tem.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Mendel M., Chen K. M., Homolka D., Gos P., Pandey R. R., McCarthy A. A., et al. (2018). Methylation of structured RNA by the m(6)A Writer METTL16 is essential for mouse embryonic development. Mol. Cell 71 986–1000.e11. 10.1016/j.molcel.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5′ UTR m(6)A promotes cap-independent translation. Cell 163 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad H. P., Barbash O., Creasy C. L. (2019). Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat. Med. 25 403–418. 10.1038/s41591-019-0376-8 [DOI] [PubMed] [Google Scholar]
- Moindrot B., Cerase A., Coker H., Masui O., Grijzenhout A., Pintacuda G., et al. (2015). A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 12 562–572. 10.1016/j.celrep.2015.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M. S., Lauritsen J., Kier M. G., Bandak M., Appelt A. L., Agerbaek M., et al. (2016). Late relapses in stage I testicular cancer patients on surveillance. Eur. Urol. 70 365–371. 10.1016/j.eururo.2016.03.016 [DOI] [PubMed] [Google Scholar]
- Muller S., Bley N., Glass M., Busch B., Rousseau V., Misiak D., et al. (2018). IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 46 6285–6303. 10.1093/nar/gky229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Yao S., Zhou Y., Liu Y., Huang P., Zhou A., et al. (2019). Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer 18:143. 10.1186/s12943-019-1079-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Lin Z., Wan A., Chen H., Liang H., Sun L., et al. (2019). RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer 18:46. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. (2013). N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 38 204–209. 10.1016/j.tibs.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerdoss S., Eedunuri V. K., Yadav P., Timilsina S., Rajamanickam S., Viswanadhapalli S., et al. (2018). Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 4:eaar8263. 10.1126/sciadv.aar8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D. P., Chen C. K., Pickering B. F., Chow A., Jackson C., Guttman M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton K. E., Chen B., Liu K., Hunter O. V., Xie Y., Tu B. P., et al. (2017). The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 169 824–835.e14. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W., Li J., Chen R., Gu Q., Yang P., Qian W., et al. (2019). Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 38:393. 10.1186/s13046-019-1408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Friderici K., Rottman F. (1975). The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell 4 387–394. 10.1016/0092-8674(75)90159-2 [DOI] [PubMed] [Google Scholar]
- Pi J., Wang W., Ji M., Wang X., Wei X., Jin J., et al. (2020). YTHDF1 promotes gastric carcinogenesis by controlling translation of FZD7. Cancer Res. 10.1158/0008-5472.CAN-20-0066 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Ping X. L., Sun B. F., Wang L., Xiao W., Yang X., Wang W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini B. I., Campbell S. C., Escudier B. (2009). Renal cell carcinoma. Lancet 373 1119–1132. 10.1016/S0140-6736(09)60229-4 [DOI] [PubMed] [Google Scholar]
- Roundtree I. A., Evans M. E., Pan T., He C. (2017a). Dynamic RNA modifications in gene expression regulation. Cell 169 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I. A., Luo G. Z., Zhang Z., Wang X., Zhou T., Cui Y., et al. (2017b). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 6:e31311. 10.7554/eLife.31311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salta E., De Strooper B. (2017). Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 18 627–640. 10.1038/nrn.2017.90 [DOI] [PubMed] [Google Scholar]
- Schwartz S., Mumbach M. R., Jovanovic M., Wang T., Maciag K., Bushkin G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8 284–296. 10.1016/j.celrep.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servaes S. E., Hoffer F. A., Smith E. A., Khanna G. (2019). Imaging of Wilms tumor: an update. Pediatr. Radiol. 49 1441–1452. 10.1007/s00247-019-04423-3 [DOI] [PubMed] [Google Scholar]
- Shen C., Xuan B., Yan T., Ma Y., Xu P., Tian X., et al. (2020). m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer 19:72. 10.1186/s12943-020-01190-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., Zhao B. S., Ma H., Hsu P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27 315–328. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Zheng C., Jin Y., Bao B., Wang D., Hou K., et al. (2020). Reduced Expression of METTL3 Promotes Metastasis of Triple-Negative Breast Cancer by m6A Methylation-Mediated COL3A1 Up-Regulation. Front. Oncol. 10:1126. 10.3389/fonc.2020.01126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., et al. (2017). S-Adenosylmethionine Synthesis Is Regulated by Selective N(6)-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 21 3354–3363. 10.1016/j.celrep.2017.11.092 [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70 7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- Strick A., von Hagen F., Gundert L., Klumper N., Tolkach Y., Schmidt D., et al. (2020). The N(6) -methyladenosine (m(6) A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 125 617–624. 10.1111/bju.15019 [DOI] [PubMed] [Google Scholar]
- Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., et al. (2018). R-2HG Exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172 90–105.e23. 10.1016/j.cell.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe A., Tanikawa K., Tsunetomi M., Takai K., Ikeda H., Konno J., et al. (2016). RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1alpha mRNA is translated. Cancer Lett. 376 34–42. 10.1016/j.canlet.2016.02.022 [DOI] [PubMed] [Google Scholar]
- Tang B., Yang Y., Kang M., Wang Y., Wang Y., Bi Y., et al. (2020). m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 19:3. 10.1186/s12943-019-1128-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 115 E325–E333. 10.1073/pnas.1717794115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan A., Patil V., Arora A., Hegde A. S., Arivazhagan A., Santosh V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37 522–533. 10.1038/onc.2017.351 [DOI] [PubMed] [Google Scholar]
- Wang H., Deng Q., Lv Z., Ling Y., Hou X., Chen Z., et al. (2019). N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol. Cancer 18:181. 10.1186/s12943-019-1108-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Q., Chen C., Ding Q., Zhao Y., Wang Z., Chen J., et al. (2020). METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69 1193–1205. 10.1136/gutjnl-2019-319639 [DOI] [PubMed] [Google Scholar]
- Wang S., Chim B., Su Y., Khil P., Wong M., Wang X., et al. (2019). Enhancement of LIN28B-induced hematopoietic reprogramming by IGF2BP3. Genes Dev. 33 1048–1068. 10.1101/gad.325100.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 575–578. 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- Wang Y., Gao M., Zhu F., Li X., Yang Y., Yan Q., et al. (2020). METTL3 is essential for postnatal development of brown adipose tissue and energy expenditure in mice. Nat. Commun. 11:1648. 10.1038/s41467-020-15488-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Kong S., Tao M., Ju S. (2020). The potential role of RNA N6 methyladenosine in Cancer progression. Mol. Cancer 19:88. 10.1186/s12943-020-01204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z., Zhao J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warda A. S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Hobartner C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18 2004–2014. 10.15252/embr.201744940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Lv R., Ma H., Shen H., He C., Wang J., et al. (2018). Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 69 1028–1038.e6. 10.1016/j.molcel.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Pan X., Yu Y., Yang B. (2020). Down-regulation of FTO promotes proliferation and migration, and protects bladder cancer cells from cisplatin-induced cytotoxicity. BMC Urol. 20:39. 10.1186/s12894-020-00612-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H., Huang H., Wu H., Qin X., Zhao B. S., Dong L., et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 22 191. 10.1016/j.stem.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. B., Horton J. R., Zhou J., Bedford M. T., Blumenthal R. M., Zhang X., et al. (2020). Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA. Nucleic Acids Res. 10.1093/nar/gkaa604 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S., Liang X., Yin S., Liu J., Xiang Z. (2020). N6-methyladenosine methyltransferase METTL3 promotes colorectal cancer cell proliferation through enhancing MYC expression. Am. J. Transl. Res. 12 1789–1806. [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Li X., Mu Z., Zhou J., Zhou P., Xie C., et al. (2020). FTO inhibition enhances the anti-tumor effect of temozolomide by targeting MYC-miR-155/23a cluster-MXI1 feedback circuit in glioma. Cancer Res. 10.1158/0008-5472.CAN-20-0132 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Xie H., Li J., Ying Y., Yan H., Jin K., Ma X., et al. (2020). METTL3/YTHDF2 m(6) A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J. Cell. Mol. Med. 24 4092–4104. 10.1111/jcmm.15063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang H., Zhao W., Fu S., Li Y., Ni W., et al. (2020). SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics 10 5671–5686. 10.7150/thno.42539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Yang Y., Feng G. H., Sun B. F., Chen J. Q., Li Y. F., et al. (2017). Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 27 1100–1114. 10.1038/cr.2017.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yu Y., Zong K., Lv P., Gu Y. (2019). Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 38:497. 10.1186/s13046-019-1470-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Jin H., Que B., Chao Y., Zhang H., Ying X., et al. (2019). Dynamic m(6)A mRNA methylation reveals the role of METTL3-m(6)A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene 38 4755–4772. 10.1038/s41388-019-0755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wei J., Cui Y. H., Park G., Shah P., Deng Y., et al. (2019). m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10:2782. 10.1038/s41467-019-10669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang S., He C., Xue P., Zhang L., He Z., et al. (2020). METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 19:46. 10.1186/s12943-020-1146-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Li J., Feng G., Gao S., Wang Y., Zhang S., et al. (2017). MicroRNA-145 Modulates N(6)-Methyladenosine Levels by Targeting the 3′-Untranslated mRNA Region of the N(6)-Methyladenosine Binding YTH Domain Family 2 Protein. J. Biol. Chem. 292 3614–3623. 10.1074/jbc.M116.749689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R., Li Q., Feng Z., Cai L., Xu Q. (2019). m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int. J. Mol. Sci. 20:1323. 10.3390/ijms20061323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Du Y., Wang L., Liu X. (2020). The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J. Cancer 11 3588–3595. 10.7150/jca.42338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue B., Song C., Yang L., Cui R., Cheng X., Zhang Z., et al. (2019). METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol. Cancer 18:142. 10.1186/s12943-019-1065-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Jaffrey S. R. (2020). A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 181 1582.e18–1595.e18. 10.1016/j.cell.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Huang S., Zhuang H., Ruan S., Zhou Z., Huang K., et al. (2020). YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 39 4507–4518. 10.1038/s41388-020-1303-7 [DOI] [PubMed] [Google Scholar]
- Zhang F., Xiang S., Cao Y., Li M., Ma Q., Liang H., et al. (2017). EIF3D promotes gallbladder cancer development by stabilizing GRK2 kinase and activating PI3K-AKT signaling pathway. Cell Death Dis. 8:e2868. 10.1038/cddis.2017.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., et al. (2015). N6-methyladenine DNA modification in Drosophila. Cell 161 893–906. 10.1016/j.cell.2015.04.018 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang X., Zhang X., Wang J., Ma Y., Zhang L., et al. (2019). RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U.S.A. 116 976–981. 10.1073/pnas.1812536116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Cui Y., Liu L., Ma X., Qi X., Wang Y., et al. (2020). METTL3 Facilitates Oral Squamous Cell Carcinoma Tumorigenesis by Enhancing c-Myc Stability via YTHDF1-Mediated m(6)A Modification. Mol. Ther. Nucleic Acids 20 1–12. 10.1016/j.omtn.2020.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yang Y., Sun B. F., Shi Y., Yang X., Xiao W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24 1403–1419. 10.1038/cr.2014.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Tao Z., Chen X. (2020). Identification of a three-m6A related gene risk score model as a potential prognostic biomarker in clear cell renal cell carcinoma. PeerJ 8:e8827. 10.7717/peerj.8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Liu C., Xu L., Yuan Y., Zhao J., Zhao W., et al. (2020). N(6) -methyladenosine Reader Protein Ythdc2 Suppresses Liver Steatosis via Regulation of mRNA Stability of Lipogenic Genes. Hepatology. 10.1002/hep.31220 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., Zhang X., Jaffrey S. R., Qian S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526 591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Bai Z. L., Xia D., Zhao Z. J., Zhao R., Wang Y. Y., et al. (2018). FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting beta-catenin through mRNA demethylation. Mol. Carcinog. 57 590–597. 10.1002/mc.22782 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Huang T., Siu H. L., Wong C. C., Dong Y., Wu F., et al. (2017). IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol. Cancer 16:77. 10.1186/s12943-017-0647-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Beaulaurier J., Deikus G., Wu T. P., Strahl M., Hao Z., et al. (2018). Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 28 1067–1078. 10.1101/gr.231068.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Si Y., Xu J., Lin Y., Wang J. Z., Cao M., et al. (2020). Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J. Cell. Mol. Med. 24 3521–3533. 10.1111/jcmm.15042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang C., Zhuang C., Luo X., Huang X., Yao L., Li J., et al. (2019). N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1alpha signalling axis. J. Cell. Mol. Med. 23 2163–2173. 10.1111/jcmm.14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang M., Li X., Zhu J., Zhang J., Niu F., Liang F., et al. (2019). The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 47 4765–4777. 10.1093/nar/gkz157.reA [DOI] [PMC free article] [PubMed] [Google Scholar]