Abstract
Three novel fluorescent biosensors sensitive to terahertz (THz) radiation were developed via transformation of Escherichia coli (E. coli) cells with plasmids, in which a promotor of genes matA, safA, or chbB controls the expression of a fluorescent protein. The biosensors were exposed to THz radiation from two sources: a high-intensity pulsed short-wave free electron laser and a low-intensity continuous long-wave IMPATT-diode-based device. The threshold and dynamics of fluorescence were found to depend on radiation parameters and exposure time. Heat shock or chemical stress yielded the absence of fluorescence induction. The biosensors are evaluated to be suitable for studying influence of THz radiation on the activity of gene networks related with considered gene promoters.
1. Introduction
The research field of electromagnetic radiation (EMR) of the THz frequency range has been rapidly advancing in the last three decades. Overlapping the frequencies from 0.1 to 10 THz (1 THz = 1012 hertz), corresponding to the wavelength interval of 30 µm – 3 mm [1], this spectral band attracts considerable interest both in fundamental and applied sciences owing to unique potentialities of THz waves unachievable for adjoining infrared and microwave domains. The development and perfecting of sources and detectors of THz radiation contribute to the expansion of its practical applications. At present, the variety of these applications, both potential and already implemented ones, is vast: from molecular spectroscopy and nondestructive testing to scanning security systems and medical imaging [2–6].
In parallel with the introduction of THz technologies into a human life, there are studies on the influence of THz EMR on living objects. Since in natural environment THz waves, which come mostly from the sun, are strongly absorbed by atmospheric gases [7,8], such EMR is rare in the habitats of biological organisms in the Earth biosphere. Accordingly, technogenic-origin THz waves of relatively high intensity do not have the corresponding evolutionarily caused adaptive reactions in living organisms including humans. This situation aggravates the problem of the impact of the THz factor on living systems.
Data on non-thermal effects of THz EMR on living objects have been obtained in studies on various biological entities: biomolecules, cells, tissues, and multicellular organisms [9–13]. In such studies, a wide variety of biological objects under study, THz exposure conditions, and analytical methods were presented. Considering this diversity, various results were obtained, up to the absence of any changes in the studied biological characteristics [14–16]. It has been shown that THz EMR affects the cellular genetic apparatus: both the structure and integrity of the genome [17–19] and the expression of genes [20–26]. THz irradiation causes patterns of differential gene expression that have a unique feature [20–24,26], and this phenomenon underscores the specificity of the living system response to this physical factor. These observations point to a substantial influence of THz radiation on biological systems and make highly relevant the issue of biosafety for the currently introduced THz technologies.
It is worth mentioning that, when investigating the non-thermal effects of THz EMR on gene activity, the use of fluorescent biosensors enables observation of time-dependent expression in live cells for certain genes whose activity is subject to the influence of this radiation. This study is aimed at the development and testing of E. coli-cell-based fluorescent microbial biosensors, each of which is a genetically engineered microorganism with an artificial genetic construct, i.e. a plasmid that combines two components: an inducible promoter and a reporter gene. The former is a promoter that is activated by THz irradiation of cells, and the latter is a structural gene that is regulated by this promoter in the construct and encodes a detectable fluorescent protein.
The principle of this biosensor technology is actively exploited in other fields. In particular, similar biosensors have been designed for quantitation of various toxic substances in a medium [27–30]. Our research group has previously created two THz-sensitive E. coli-based biosensors, in which promoters of catalase gene kagG and copper chaperone gene copA serve as inducible promoters [25,31,32]. The induction of these biosensors reflects a targeted influence of THz radiation on the genetic networks responsive to oxidative stress and copper ion homeostasis. For in-depth and comprehensive research into the impact of THz EMR on the gene expression apparatus, it is necessary to create a series of highly sensitive biosensors in which activation of various specific inducible promoters (and therefore the cellular fluorescent response) reflects the state of certain genetic (metabolic) networks.
In this study, to create THz-sensitive recombinant genetic constructs, we selected the promoters that participate in regulation of transcription factor (TF) genes. Such genes encode regulatory proteins, every of which is able to control a group of genes (i.e. a regulon) as part of one or several operons [33]. Consequently, the induction of each newly developed biosensor caused by THz irradiation or another stimulus may represent a specific pattern of activities for genes corresponding to some TFs and its target genes. In total, we utilized three inducible promoters and two genes of fluorescent proteins to design THz-sensitive biosensors (E. coli/pMatA-TurboGFP, E. coli/pSafA-TurboGFP, and E. coli/pChbB-TurboYFP) and investigated their induction.
2. Materials and methods
2.1. Construction of biosensor plasmids
The construction of recombinant biosensor plasmids pMatA-TurboGFP, pSafA-TurboGFP, and pChbB-TurboYFP was carried out in two stages: preparation of the necessary DNA fragments and their joining by the Gibson method [34]. As basic vectors, we used plasmids pTurboGFP-B [35] and pTurboYFP-B [36] (Evrogen, Russia), carrying respectively a gene of fluorescent protein TurboGFP or TurboYFP, under the control of the T5 promoter/lac operator element (this element consists of the phage T5 promoter and two lac operator sequences). Both vectors contain a gene of ampicillin resistance. Proteins TurboGFP [37] and TurboYFP [38] respectively are enhanced variants of green fluorescent protein CopGFP from the copepod Pontellina plumata and yellow fluorescent protein PhiYFP from the jelly fish Phialidium sp. [39].
DNA fragments containing a gene of a structural protein, TurboGFP or TurboYFP, were amplified from basic vectors pTurboGFP-B and pTurboYFP-B by polymerase chain reaction (PCR) with specific primers (Table). The reactions were carried out by means of the High Fidelity PCR Kit (New England Biolabs, UK) in a final volume of 30 µL, including 1 µL of the vector (pTurboGFP-B or pTurboYFP-B) diluted 1000-fold, 6 µL of 5X Q5 Reaction Buffer Pack (at the final concentration, it contains 2 mM MgCl2), 6 µL of 5X Q5 High GC Enhancer, 0.6 µL each of 10 mM dNTPs, 1 µL each of 10 µM primers, 12.3 µL of water, and 0.3 µL of Q5 Hot Start High-Fidelity DNA Polymerase (2 U/µL). The amplification program was as follows: initial denaturation of the template for 2 min at 95°C; followed by 33 cycles of denaturing for 15 s at 95°C, annealing for 15 s at 60°C, and extension for 4 min at 72°C; with final extension at 72°C for 5 min. After PCR, the reaction mixture was purified (to remove the original plasmid) by the addition of 2 µL of the MalI restriction enzyme (SibEnzyme, Russia) with incubation at 37°С for 30 min. As a result, DNA fragments were obtained that — in contrast to the basic vectors — did not contain only a region of the T5 promoter/lac operator element.
DNA fragments containing the matA, safA, or chbB promoter were derived from genomic DNA of E. coli cells of the JM109 strain by PCR with specific primers (Table 1). By means of the diaGene Kit for Isolation of Genomic DNA from Bacterial Cells (Dia-M, Russia; according to the manufacturer’s protocol), the genomic DNA was first isolated from bacterial cells grown overnight (37°C, 250 rpm) in the Lysogeny broth (LB) medium (1% (w/v) of tryptone, 0.5% (w/v) of yeast extract, and 171 mM NaCl, pH 8.0) containing 30 µg/mL nalidixic acid. The thermal conditions of PCR were similar to those for the previous stage, except for minor modifications: as a template, we utilized 1 µL of an undiluted genomic-DNA sample, and in the cycling program, the extension stage was 2 min long. In this way, DNA fragments were obtained containing 5′ overhangs complementary to the 5′ ends of one of the DNA fragments containing the gene of the TurboGFP or TurboYFP protein.
Table 1. Primers used for PCR.
| DNA template, length (bp) | Primer sequences (5′→3′) | Obtained PCR product, length (bp) |
|---|---|---|
| Vector pTurboGFP-B, 4102 | Forward: gagagcgacgagagcg Reverse: tttctcgaggtgaagacgaaagg | Fragment of vector pTurboGFP-B, 3980 |
| Vector pTurboYFP-B, 4140 | Forward: atgagcagcggcgcc Reverse: tttctcgaggtgaagacgaaagg | Fragment of vector pTurboYFP-B, 4017 |
| E. coli JM109 genomic DNA | Forward: cctttcgtcttcacctcgagaaatgccaataaccttatgaaaccaaatgtc Reverse: cgctctcgtcgctctctactttccaaacctgtaatttttacaggtg | Fragment of genomic DNA containing matA promoter, 307 |
| Forward: cctttcgtcttcacctcgagaaatcatccgcacagcagaagaattc Reverse: cgctctcgtcgctctcttcaaatatgtttatttagcggataacgttaaaaat | Fragment of genomic DNA containing safA promoter, 262 | |
| Forward: cctttcgtcttcacctcgagaaatcaattcgtctgaaagcttgagtaac Reverse: ggcgccgctgctcataaaactgccctcatcgacga | Fragment of genomic DNA containing chbB promoter, 336 |
The length of the resultant DNA fragments was verified by 1% agarose gel electrophoresis with ethidium bromide staining.
Next, the obtained DNA fragments were joined by the Gibson method via the kit Gibson Assembly Master Mix (New England Biolabs, UK): the fragment of pTurboGFP-B was fused either with the DNA fragment containing the matA gene promoter or with the DNA fragment containing the safA gene promoter; the pTurboYFP-B fragment was fused with the DNA fragment containing the chbB gene promoter. For this purpose, on ice, we prepared a mixture containing 1 µL each of the DNA fragments being fused and 2 µL of 2X Gibson Assembly Master Mix. This mixture was incubated for 15 min at 50°С.
2.2. Creation of the biosensor cells
To create the bacterial biosensor cells, E. coli was transformed with the biosensor plasmids by electroporation.
First, electrocompetent E. coli JM109 cells were prepared. To this end, bacterial cells grown overnight (37°С, 250 rpm, in the LB medium containing 30 µg/mL nalidixic acid) were seeded in 200 mL of the fresh antibiotic-free LB medium and were incubated under the same conditions until OD600 (optical density at 600 nm wavelength, assuming 1 cm path length) reached 0.6. After that, the cells were cooled to 4°С and washed twice with water and then once with 10% aqueous glycerol (with pelleting by centrifugation for 10 min at 2200 × g and 4°С, followed by resuspension). After the washes, the cells were resuspended in 1 mL of 10% aqueous glycerol; the suspension was divided into 40 µL aliquots and frozen in liquid nitrogen for subsequent use.
Second, the bacterial-cell transformation was performed. For this purpose, to a thawed aliquot of the electrocompetent cells, we added 1 µL of an obtained biosensor plasmid (pMatA-TurboGFP, pSafA-TurboGFP, or pChbB-TurboYFP), and electroporation was conducted in the Gene Pulser Cuvette with a gap width of 1 mm (Bio-Rad, USA) on Gene Pulser Xcell (Bio-Rad) via the preset protocol for bacterial cells (voltage 1.8 kV, capacity 25 µF, and resistance 200 Ω). After the electroporation, 0.5 mL of Super Optimal broth with catabolite repression (SOC medium: 2% (w/v) of tryptone, 0.5% (w/v) of yeast extract, 20 mM glucose, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, and 10 mM MgSO4, pH 7.5) was added to the cells, and the mixture was incubated for 1 h at 37°C. Next, it was transferred to Petri dishes with an LB agar medium (the LB medium supplemented with 1.5% (w/v) agar agar) containing 100 µg/mL ampicillin.
At the third stage, the selected clones were confirmed by sequencing. To this end, cells of colonies from the Petri dishes were transferred onto the LB medium containing 100 µg/mL ampicillin, were grown overnight (37°С, 250 rpm), and were subjected to plasmid DNA isolation by means of the diaGene Kit for Isolation of Plasmid DNA (Dia-M, Russia) according to the manufacturer’s protocol. The DNA sequence on the border between the inducible promoter (matA, safA or chbB) and the gene of a fluorescent structural protein (TurboGFP or TurboYFP) was amplified by PCR and analyzed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol, on an ABI Prism 3100 Avant Genetic Analyzer (Applied Biosystems, USA). After the verification of the DNA sequences in the recombinant constructs, an overnight culture of the biosensor cells was transferred to 10% aqueous glycerol via pelleting by centrifugation (for 10 min at 2200 × g) with subsequent resuspension; the suspensions was divided into 40 µL aliquots and frozen in liquid nitrogen for subsequent use.
2.3. Exposure of the biosensor cells to THz radiation
From the frozen aliquots, biosensor cells were seeded on the LB medium containing 100 µg/mL ampicillin, were grown overnight (37°С, 250 rpm), reseeded on an identical fresh medium with the antibiotic, and were grown until OD600 = 0.5 for ∼2.5 h. Then, via pelleting by centrifugation (for 3 min at 6000 × g) with subsequent resuspension, the cells were washed to remove the old medium and were transferred onto a fresh medium (containing 100 µg/mL ampicillin), where they were irradiated and analyzed.
Irradiation experiments were conducted separately with two types of coherent THz radiation sources: pulsed short-wave and continuous long-wave.
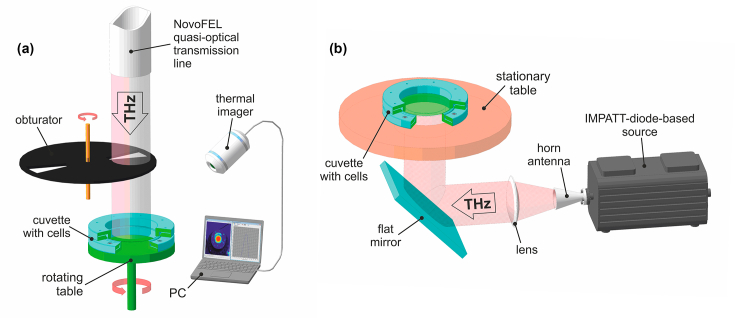
In the former case, the radiation source was the Novosibirsk Free Electron Laser (NovoFEL) at the Shared Research Facility “Siberian Synchrotron and Terahertz Radiation Centre” (Budker Institute of Nuclear Physics of the Siberian Branch of the Russian Academy of Sciences (SB RAS), Novosibirsk, Russia). The part of NovoFEL that is called “the first stage” was used; it provides pulsed THz radiation with general characteristics described elsewhere [40]. For this work, a specially equipped biological workstation was employed; it allows an investigator to control and regulate the irradiation dose (Fig. 1(a)). The irradiation of cultured biosensor cells (50 µL) was performed in a specially designed 60-mm-diameter cuvette that was made of two THz-transparent 40-µm-thick polypropylene membranes stretched onto metallic rings. The sample in the cuvette assumed the shape of a 40-mm-diameter 40-µm-thick circular horizontal layer. The diameter (full width at half maximum of intensity) of the Gaussian THz beam in the plane of the sample was ∼30 mm (σ ≈ 13 mm). The horizontally positioned cuvette was irradiated from above, and to ensure radially uniform exposure, the cuvette was rotated on a special rotating table. Due to relatively high average power density of this THz radiation, samples could get heated during exposure, and this process was controlled by a TKVr-SVIT101 high-precision thermal imager (Rzhanov Institute of Semiconductor Physics of the Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia) with 0.03°C accuracy [41]. The average radiation power and consequent heating of the samples were regulated with an electrically-driven obturator consisting of two rotating closely spaced copper disks (25 cm in diameter), which had two sector apertures and were mounted on a common rotational axis. The average power density of THz radiation in the plane of the sample was measured with an IMO-4 detector (experimental factory Etalon; Volgograd, Russia). The THz exposure conditions were as follows: radiation frequency, 2.31 THz; pulse repetition rate, 5.6 MHz; pulse duration, 100 ps; average power density, ∼140 mW/cm2; temperature of samples upon heating, 36°С ±1°С; exposure time, 15 or 30 min. In parallel, control cultured cells were incubated in an identical cuvette in an air thermostat at 37°С for the same period.
Fig. 1.
General schemes of experiments with irradiating cultured-bacterial-cells samples by the NovoFEL (a) and TeraSense source (b).
The second source of THz radiation was a compact solid-state device based on an IMPATT-diode (Terasense Group Inc., USA), which ensured coherent continuous-wave emission of THz EMR with a frequency of 0.14 THz and output power of 44 mW (Fig. 2(b)) [42]. In this setup, the biosensor cells were irradiated in a cuvette similar to the one used above. The radiation was emitted from the source into free space through a matched horn-lens antenna as a collimated (plane) wave beam propagating in a horizontal direction. After that, the THz beam was deflected in a vertical direction by a flat mirror and hit the horizontally positioned immobile cuvette with the sample by irradiating it from below. With the help of a pyroelectric detector (Microtech Instruments Inc., USA) and a quasi-optical power meter (Absolute THz Power-Energy Meter; Thomas Keating Ltd., UK), we measured the diameter (full width at half maximum of intensity) of the quasi-Gaussian radiation beam and the average power density in the plane of the sample, which were found to be ∼30 mm and ∼2 mW/cm2, respectively. Cultured biosensor cells, just as in the first case, were irradiated in a 50 µL volume for 15 or 30 min under ambient conditions (∼26°С). Via measurements with a calibrated thermocouple, it was determined that the heating of the sample during the THz exposure was negligible and did not exceed 1°С. In parallel, control cultured cells were incubated in an identical cuvette at room temperature for the same period.
Fig. 2.
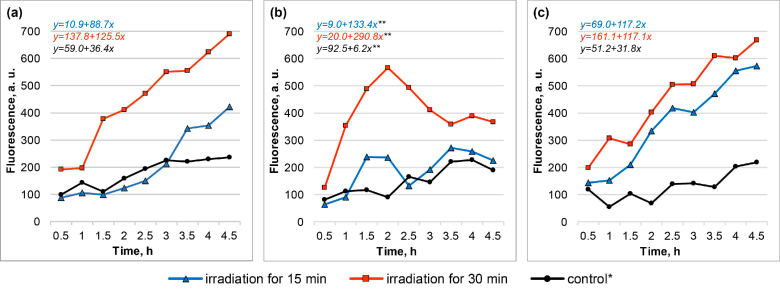
Fluorescence dynamics (fluorescence curves and the respective linear regression equations) of biosensor cells in response to THz irradiation by NovoFEL: E. coli/pMatA-TurboGFP (a), E. coli/pSafA-TurboGFP (b), and E. coli/pChbB-TurboYFP (c), in comparison with the control (bulk heating). Results of one independent replication are presented. *Incubation of cells in the cuvette at 37°С for 30 min. **Equation corresponds to a period up to a 2 h time point only.
After the irradiation in both cases (pulsed and continuous-wave), the samples were transferred into a 96-well flat-bottom polystyrene microplate (Costar-3599, Corning, USA), and fluorescence was measured on a VICTOR X3 2030 microplate reader (Perkin Elmer, USA) at the following settings: excitation wavelength, 485 nm; emission wavelength, 535 nm; duration of irradiation by exciting light, 0.1 s. The measurements were carried out for 4.5 h after the end of THz exposure, with signal acquisition at 30 min intervals; during the intervals, the cell samples were incubated at 37°С and 800 rpm. In case of the irradiation by NovoFEL, the first measurement was carried out 30 minutes after the end of the irradiation.
Aside from assays of the fluorescence of proteins TurboGFP and TurboYFP in the biosensor cells, sham experiments were conducted: we analyzed the fluorescence of these proteins in E. coli cells bearing a plasmid in which structural gene TurboGFP or TurboYFP is associated with a THz-insensitive regulatory promoter. For this purpose, under identical conditions of irradiation and fluorescence quantitation, we analyzed E. coli JM109 cells transformed with basic vector pTurboGFP-B or pTurboYFP-B, in which the genes of proteins TurboGFP and TurboYFP are regulated by the T5 promoter/lac operator element, and which also confer ampicillin resistance.
2.4. Exposure of the biosensor cells to an elevated temperature or various chemical stressors
To evaluate the specificity of induction of the newly developed biosensors, we conducted experiments with thermal or chemical stress. To this end, biosensor cells were subjected to the same analytical procedures as in the THz irradiation protocol.
During the heat shock experiment, cell cultures were dispensed into wells of a 96-well microplate at 50 µL/well. The microplates with experimental and control samples were incubated for 30 min in the air thermostat at 42°C and 37°С, respectively.
In the assays of chemical inducers (namely, hydrogen peroxide, phenol, salicylic acid, Cu(II) sulfate, or Fe(III) chloride), in the preparation procedure, during the transfer onto a fresh LB medium, the cell cultures were concentrated 1.11-fold. The cultures were dispensed into wells of a 96-well microplate at 45 µL/well; next, the experimental and control samples were mixed with 5 µL of the LB medium with a chemical inducer or without it, respectively. The inducers were applied in the following ranges of final concentrations (serial twofold dilutions): 0.156–40 mM for hydrogen peroxide and phenol, 0.005–1.25 mM for salicylic acid, and 0.6–156 µМ for Cu(II) sulfate and Fe(III) chloride.
Immediately after the thermal or chemical stress, the fluorescence was measured similarly to the THz experiments.
2.5. Statistical analysis
Every type of experiment with THz irradiation and the experiments with the elevated temperature and chemical stressors were implemented in five independent replications. Data from the fluorescence curves were subjected to linear regression analysis. For each paired group (inside each replication), slope coefficients were calculated in an experiment and control, and significance of their differences according to the sum of all independent replications was evaluated by nonparametric Wilcoxon’s rank sum test. To plot averaged normalized induction levels, in each paired group (inside each replication), we computed a ratio of fluorescence intensity in an experiment to that in the control at a given time point and then averaged all five independent replications. All the calculations were carried out in Statistica 10 software (StatSoft, USA).
3. Results
Previously, after exposing E. coli JM109 cells to the NovoFEL radiation, using genome-wide RNA sequencing screening, we identified seven TF genes that are significantly overexpressed (log2 fold change ≥1, P < 0.05) under THz impact: cadC, caiF, chbR, gadW, matA, tdcR, and ydeO [43]. In this case, the conditions of THz exposure were similar to those used in the present work, in case of the 15-minute irradiation by NovoFEL. The THz sensitive biosensors were developed that allow to monitor the activity of three genes from these seven overexpressed, i.e. matA, ydeO, and chbR; after the RNA sequencing screening, they manifested a log2 fold change of 1.31, 3.11, and 3.03, respectively [43]. The activity of these TF genes in the E. coli genome is regulated by the following promoters: pmatA [44], psafA (as part of the safA-ydeO operon) [45], and pchbB (as part of the chbBCARFG operon) [46]. Accordingly, during the assembly of the biosensor genetic constructs, these promoters were chosen as inducible ones.
Thus, recombinant DNA constructs pMatA-TurboGFP, pSafA-TurboGFP, and pChbB-TurboYFP were obtained in which the promoter of matA, safA, or chbB control the expression of a fluorescent protein: TurboGFP or TurboYFP. By transformation of E. coli JM109 cells with the plasmids, biosensors E. coli/pMatA-TurboGFP, E. coli/pSafA-TurboGFP, and E. coli/pChbB-TurboYFP were created, and their induction was tested under various conditions: THz irradiation, heat shock, or chemical stress.
The irradiation experiments were conducted using the NovoFEL and the compact TeraSense source at the fixed radiation parameters described above. In both cases, the THz irradiation was performed for 15 or 30 min in identical cuvettes with equal volumes of a cell culture sample and identical cell concentrations at the beginning of the irradiation. The biosensors displayed induction upon THz exposure, resulting in the production of a fluorescent protein by the cells. Nonetheless, the induction was not always detectable and showed different dynamics among the various conditions. Note, since nutrient medium LB exhibits intrinsic fluorescence at the optical detection parameters being used, all the presented data and their comparative analysis are given after subtracting the background fluorescence of the nutrient medium.
Under the impact of the high-intensity pulsed short-wave THz radiation from the NovoFEL source, the induction of the biosensors was seen both after 15 and 30 min irradiation, but the induction was more pronounced after the longer exposure (Fig. 2). For biosensors E. coli/pMatA-TurboGFP and E. coli/pChbB-TurboYFP, the growth of fluorescence persisted during the whole period of measurements (until 4.5 h after the end of the irradiation). For biosensor E. coli /pSafA-TurboGFP, a domelike curve of fluorescence was observed, with the peak corresponding to 2 h after the irradiation.
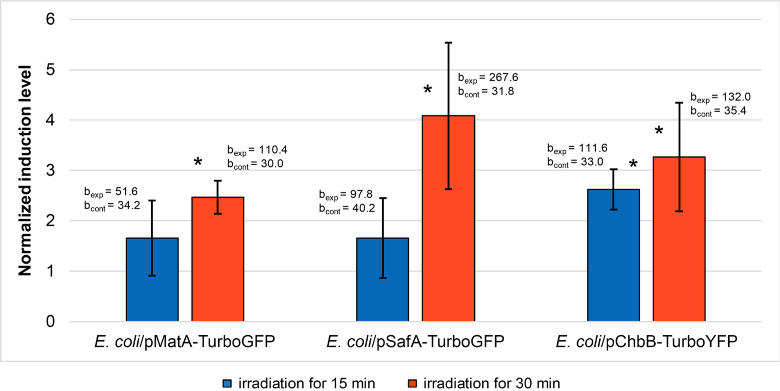
After the irradiation by NovoFEL, in each group of cells (experiment and control), average slope coefficients were calculated during the slope analysis throughout the whole period of measurements for E. coli/pMatA-TurboGFP and E. coli /pChbB-TurboYFP and only for the first 1.5 h of the measurements for E. coli/ pSafA-TurboGFP. When plotting the normalized induction levels (in irradiated and control groups), the fluorescence values corresponding to 4.5 and 2 h time points were used. The data processed in this way are presented below (Fig. 3). After 15 min irradiation, significant induction was detected only for the biosensor E. coli/pChbB-TurboYFP, whereas after 30 min irradiation it was revealed for all three biosensors.
Fig. 3.
The normalized induction levels of the biosensor cells (averaged over five independent replications) in response to THz irradiation by NovoFEL. The error bars indicate standard deviation; bexp and bcont are average slope coefficients in experiment and control, respectively. *Significant differences (P < 0.05) between slope coefficients in experiment and control.
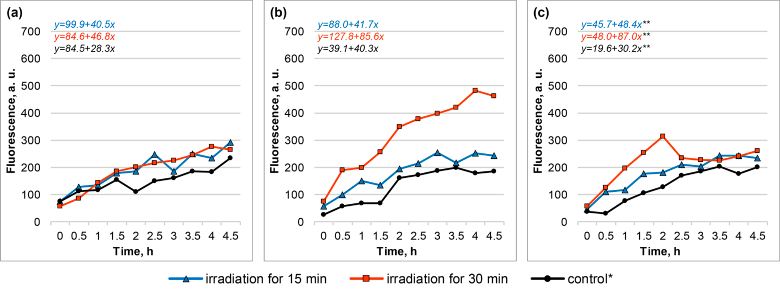
After exposure to low-intensity continuous long-wave THz radiation from the compact TeraSense source, the fluorescent response on average was substantially weaker and was qualitatively different from that after the irradiation by NovoFEL (Fig. 4). First of all, the induction of the biosensors was almost undetectable after the irradiation for 15 min. Furthermore, after the 30 min irradiation, the biosensor E. coli/pMatA-TurboGFP was not induced, while the biosensor E. coli/pSafA-TurboGFP got induced monotonously for the whole measurement period (i.e. without the peak in the middle of the period, as seen with NovoFEL), and the biosensor E. coli/pChbB-TurboYFP got induced weakly and unstably, and only during the first 1–2 h after the irradiation.
Fig. 4.
Fluorescence dynamics (fluorescence curves and the respective linear regression equations) of biosensor cells in response to THz irradiation by the TeraSense source: E. coli/pMatA-TurboGFP (a), E. coli/pSafA TurboGFP (b), and E. coli/pChbB-TurboYFP (c), as compared to the control. Results of one independent replication are presented. *Incubation of cells in the cuvette at room temperature for 30 min. **Equation corresponds to a period up to a 1 h time point only.
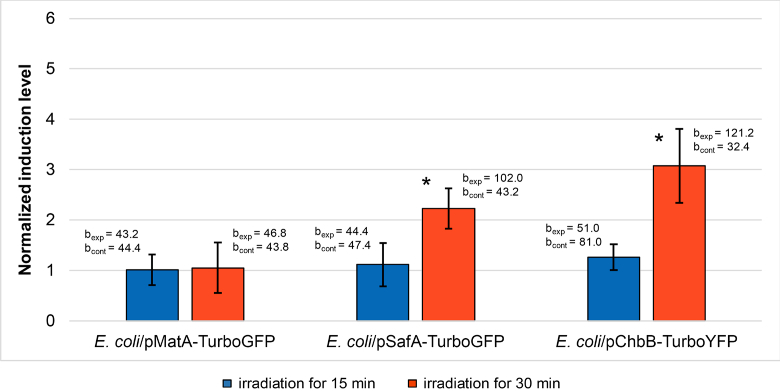
After the irradiation by TeraSense, in each cell group (experiment and control) average slope coefficients were computed during the slope analysis in the whole measurement period for E. coli/pMatA-TurboGFP and E. coli/pSafA-TurboGFP and only for the first 1 h of the measurements for E. coli/pChbB-TurboYFP. When plotting the normalized induction levels (in irradiated and control groups), the fluorescence values taken at 4.5 and 1 h time points were used. The data processed in this way are depicted below (Fig. 5). Significant induction was seen only after 30 min irradiation for the biosensors E. coli/pSafA-TurboGFP and E. coli/pChbB-TurboYFP.
Fig. 5.
Normalized induction levels of the biosensor cells (averaged over five independent replications) in response to THz irradiation by the TeraSense source. The error bars indicate standard deviation; bexp and bcont are average slope coefficients in experiment and control, respectively. *Significant differences (P < 0.05) between slope coefficients in experiment and control.
The induction of the biosensors was not observed after the biosensors’ cells were subjected to heat shock (heating at 42°C for 30 min) or a chemical stressor (hydrogen peroxide, phenol, salicylic acid, Cu(II) sulfate, or Fe(III) chloride) in all the tested concentration ranges. Examples of the fluorescence curves are presented in Supplement 1 (1.4MB, pdf) (Figs. S1 and S2); in all independent replications, the results were comparable and with no reproducible dynamics. In case of the chemical stress, the data corresponding to individual chemical inducers concentrations (in the middle of the selected range) are presented in Supplement 1 (1.4MB, pdf) (Fig. S2); at the other concentrations, the results were also negative.
There was no fluorescent response in sham experiments, i.e. after identical THz irradiation of E. coli cells harboring basic vector pTurboGFP-B or pTurboYFP-B. By the example of 30-minute irradiation, the individual fluorescence curves are presented in Supplement 1 (1.4MB, pdf) (Fig. S3).
4. Discussion
In general, this study continues our previous research [25,31,32] into the influence of THz radiation on the cellular apparatus of gene expression by means of E. coli-based fluorescent biosensors.
As we have previously demonstrated, during THz irradiation, a number of TF genes are induced, e.g. matA, ydeO, and chbR [43]. Products of the said TF genes — proteins MatA, YdeO, and ChbR — regulate various relatively specific functions in the cell (see below). The expression regulation of genes ydeO and chbR is implemented within operons safA-ydeO [45] and chbBCARFG [46], respectively, whereas matA is regulated independently [44]. Thus, the expression of proteins MatA, YdeO, and ChbR is controlled in the E. coli genome by matA, safA, and chbB promoters, respectively, which were used for the assembly of our biosensor genetic constructs.
The newly developed biosensors E. coli /pMatA-TurboGFP, E. coli/pSafA-TurboGFP, and E. coli/pChbB-TurboYFP were tested for their induction (emergence of fluorescence) when exposed to THz radiation with different parameters: high-intensity pulsed short-wave and low-intensity continuous long-wave. In both cases, a sample of biosensor cell culture was placed into a special cuvette where the sample assumed a thickness of 40 µm. It is known that THz waves are strongly absorbed by water-containing media [47]. For instance, a 100-µm-thick water layer absorbs more than 98% of radiation power at the frequency of 2.31 THz [48]. Here, the 40 µm thickness for water-based samples of cultured biosensor cells ensured their irradiation throughout the whole volume.
In this study, it has been successfully demonstrated the radiation-stimulated induction of biosensors which varied upon THz exposure conditions. Additional assays have been also performed. First, after identical THz irradiation of E. coli cells, harboring basic vector pTurboGFP-B or pTurboYFP-B (devoid of matA, safA, and chbB promoters), we proved the absence of a similar fluorescent response in Supplement 1 (1.4MB, pdf) (Fig. S3). This finding confirms that it is promoters of matA, safA, and chbB that are activated by the irradiation of the biosensor cells. Second, after the heat shock or chemical stress, the induction of the biosensors was undetectable in Supplement 1 (1.4MB, pdf) (Figs. S1 and S2). Thus, in comparison with these impacts, induction specificity of the newly developed biosensors to THz radiation was presented.
After irradiating the biosensors at NovoFEL by short-wave pulsed 2.31 THz radiation with an average power density ∼140 mW/cm2, we observed relatively strong induction (Figs. 2 and 3). In case of 15-minute exposure, the irradiation conditions in these experiments were analogous to the ones evaluated previously during the genome-wide RNA sequencing screening. To test the obtained biosensors, it was important to accurately reproduce the former conditions of THz irradiation. In our current study we also found that after the longer exposure (30 min) the induction of all three biosensors was significant and more pronounced relative to the 15 min irradiation (Figs. 2 and 3).
When the biosensors were irradiated by continuous long-wave 0.14 THz radiation from a TeraSense source with an average power densihighty ∼2 mW/cm2, the induction was qualitatively different and weaker overall (Figs. 4 and 5). In this case, the cells were exposed to THz radiation with physical characteristics substantially different from those of NovoFEL, including a 70-fold lower average intensity. As a consequence, under these irradiation conditions, all three biosensors failed to be induced at 15 min exposure time. The biosensor E. coli/pMatA-TurboGFP was not induced even after 30 min. On the contrary, the biosensors E. coli/pSafA-TurboGFP and E. coli/pChbB-TurboYFP were induced when irradiated during 30 min, and averaged normalized induction of the former was ∼2-fold weaker than (while for the latter it was comparable to) that for 30 min exposure at NovoFEL (Figs. 4 and 5).
The results of our study allow us to highlight the following effects observed. First, the created biosensors were proven to respond to THz radiation with very different physical parameters; this response showed itself through gene expression. Second, these biosensors and, therefore, the promoters of matA, safA, and chbB diverged in sensitivity to THz exposure conditions. At the level of the E. coli genome, this phenomenon is manifested in selective expression of the respective genes, including TF genes matA, ydeO, and chbR. In our experiments, THz fluxes from alternative radiation sources we employed were noticeably different in characteristics: intensity (average power density, 140 vs. 2 mW/cm2), frequency (2.31 vs. 0.14 THz), and the operation regime (pulsed vs. continuous). Accordingly, the expression of the analyzed genes (judging by the biosensor induction) was sensitive to these parameters. This observation is consistent with the results of other studies, which revealed the importance of one or another characteristic of THz radiation for specific activation or repression of certain genes. In this way, researchers have proven the influence of experimentally varied parameters of THz radiation: intensity (for human keratinocytes and fibroblasts subjected to radiation pulses with energies of 0.1 and 1 µJ [18,22] and radiation with pulse intensities in the range of 0.6–47.2 MW/cm2 [49], frequency (for human keratinocytes under irradiation at 1.4, 2.52, and 3.11 THz [50]), and the operation regime and frequency spectrum (for mouse mesenchymal stem cells during exposure to continuous-wave single-frequency radiation at 2.52 THz and pulsed broadband radiation centered at 10 THz [20,51]). In our case, there is no definitive answer to the question which of the above-mentioned radiation parameters is of primary importance for the observed differences in the induction of the new biosensors. And finally, the third effect to be noted: both for the NovoFEL and TeraSense sources it was clearly demonstrated a dose-dependent nature of the THz-stimulated induction. For experiments with TeraSense it had the threshold behavior: the effect was not revealed at 15 min irradiation.
It is worth mentioning that regulatory genes matA, ydeO, and chbR are important genetic elements of the studied loci in the E. coli genome. Activation of these genes is associated with the induction of the newly developed biosensors. Genes matA, ydeO, and chbR are involved in various metabolic processes in bacterial cells, as clarified below.
The matA gene encodes LuxR-type transcriptional dual regulator MatA, whose production is positively autoregulated. This protein takes part in (promotes) the adaptation of planktonic E. coli to an adhesive lifestyle. On the one hand, MatA serves as a key positive regulator (acting at transcriptional and post-transcriptional levels) for the expression of matBCDEF genes, which code for structural components of Mat fimbriae and implement biofilm development [44,52]. On the other hand, MatA decreases the expression of flagellar master operon flhDC [44], which encodes TFs needed for the initiation of flagellar synthesis [44,53]. Thus, matA activation is associated with bacterial biofilm formation. The formation of biofilms per se is an adaptation process and increases cell resistance to such stressors as antimicrobial compounds, heavy metals, extreme deviations in pH, oxidative stress, and oxygen-limiting conditions [54,55]. High-intensity THz irradiation at the NovoFEL is possibly a stressor (acting for example via oxidative-stress development) for such living systems as bacterial cells, and the expression of matA is indirect evidence.
The chbR gene encodes transcriptional dual regulator ChbR, which belongs to the AraC-like family and participates in the uptake and metabolism of β-glucoside chitobiose. For example, in the presence of chitobiose, ChbR induces chbBCARFG operon transcription, whereas in the absence of the inducing sugar, it represses this transcription (because ChbR is encoded within the chbBCARFG operon itself, the production of this protein is autoregulated). In the former case, ChbR cooperates with catabolite gene activator protein (CAP), and in the latter, with the negative regulator NagC [46]. Furthermore, the chbR gene is involved in the utilization of β-glucoside cellobiose, because specific mutation of this gene leads to alteration of chbBCARFG operon activity, thereby ensuring assimilation of this substrate too (E. coli cells are normally unable to metabolize cellobiose) [56]. It is reported that the growth of E. coli cells on the LB medium is carbon limited [57], and such a nutrient limitation causes changes in the expression of several transport- and assimilation-related genes [58]. At the same time, such alterations of gene transcription may vary widely depending on other accompanying factors [58]. Under the carbon-limited conditions in our experiments, THz irradiation caused activation of the chbBCARFG operon, thereby leading to the induction of metabolic genes responsible for cellular utilization of alternative carbon sources: β-glucosides chitobiose and cellobiose. Of note, according to our data on normalized induction of the biosensor, the intensity of this process was approximately equal between the two sources after 30 min exposure. Further specific studies are necessary to elucidate the mechanisms underlying the initiation of these adaptive responses in E. coli cells.
The ydeO gene codes for multifunctional transcriptional dual regulator YdeO, which is encoded within the safA-ydeO operon and is negatively autoregulated [45,59]. This TF belongs to the AraC/XylS family and coordinates the cellular response to stress (primarily to an acidic environment) and to anaerobic conditions. For instance, YdeO is a positive regulator of operons gadE-mdtEF, gadW, and slp-dctR [60,61]. The first two are involved in the glutamate-dependent acid resistance system (GAD system) [62], whereas slp-dctR also directly participates in acid resistance [63]. Additionally, YdeO serves as a positive regulator of operons hyaABCDEF and appCBA [61], which encode hydrogenase 1 and quinone oxidase, respectively (both are involved in bacterial respiration) [64,65]. Besides, there is evidence of positive regulation of operons nhaAR and yiiS-uspD by this protein [61]. Operon nhaAR codes for a sodium-hydrogen antiporter and is responsible for cell adaptation to Na+ and alkaline pH (in the presence of Na+) [66]. Operon yiiS-uspD is a player in universal antistress defense in E. coli and is involved in the protection against oxidative stress and DNA damage [67,68]. The participation of ydeO in the development of a response to THz radiation through the regulation of operons appCBA and yiiS-uspD may imply the active oxidative stress [65,68] caused by radiation.
The induction of the biosensor E. coli/pSafA-TurboGFP was more pronounced after high-intensity THz irradiation at the NovoFEL; this effect correlates with the activation of stress gene matA and allows for the development of oxidative stress during this treatment. This conclusion is consistent with the findings of our previous biosensor studies [25,31,32], which indicate activation of catalase gene katG in E. coli cells under THz irradiation.
5. Conclusion
On the basis of promoters of matA, safA, and chbB, THz-sensitive fluorescent bacterial biosensors E. coli/pMatA-TurboGFP, E. coli/pSafA-TurboGFP, and E. coli/pChbB-TurboYFP were successfully created. The induction of these biosensors is manifested as fluorescent luminosity and respectively reflects the activity of transcription units matA, safA-ydeO, and chbBCARFG, which contain TF genes matA, ydeO, and chbR.
The biosensor induction was studied under THz irradiation with various parameters and under other conditions (thermal or chemical stress), and the results can be summarized as follows:
-
•
Within the investigated conditions, the induction was found to be specific to THz radiation, since the exposure to heat shock or various chemical stresses did not yield a fluorescent response.
-
•
The induction was dose-dependent: the effect increased with increasing irradiation duration from 15 to 30 min.
-
•
The manifestation of the induction and its qualitative traits depended on parameters of THz radiation.
Biosensors E. coli/pMatA-TurboGFP, E. coli/pSafA-TurboGFP, and E. coli/pChbB-TurboYFP may be used for intravital assessment (both qualitative and quantitative) of the activity of E. coli regulons controlled by regulatory proteins MatA, YdeO, and ChbR. These biosensors may be helpful for the research into the influence of THz radiation on a living system and in other fields that require expression analysis of the genes in question. Moreover, these biosensors are potentially applicable to the development of environmental THz radiation-monitoring systems.
Supplementary Material
Acknowledgments
The irradiation experiments were conducted on the Novosibirsk Free Electron Laser at the Siberian Synchrotron and Terahertz Radiation Centre (Budker Institute of Nuclear Physics SB RAS, Novosibirsk, Russia). The authors also acknowledge the Shared Equipment Center CKP “VTAN” (ATRC, Physics Department) of Novosibirsk State University (Novosibirsk, Russia) for the provided instrumental support. The creation of the biosensors and their testing were carried out, respectively, at the Laboratory of Molecular Biotechnologies and the Kurchatov Genomics Center of Federal research center Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia.
Funding
Ministry of Education and Science of the Russian Federation10.13039/501100003443 (0324-2019-0040-C-01, 075-15-2019-1662, АААА-А17-117030310290-5).
Disclosures
The authors declare no conflicts of interest.
See Supplement 1 (1.4MB, pdf) for supporting content.
References
- 1.Romanenko S., Begley R., Harvey A. R., Hool L., Wallace V. P., “The interaction between electromagnetic fields at megahertz, gigahertz and terahertz frequencies with cells, tissues and organisms: risks and potential,” J. R. Soc., Interface 14(137), 20170585 (2017). 10.1098/rsif.2017.0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhillon S. S., Vitiello M. S., Linfield E. H., Davies A. G., Hoffmann M. C., Booske J., Paoloni C., Gensch M., Weightman P., Williams G. P., Castro-Camus E., Cumming D. R. S., Simoens F., Escorcia-Carranza I., Grant J., Lucyszyn S., Kuwata-Gonokami M., Konishi K., Koch M., Schmuttermaer C. A., Cocker T. L., Huber R., Markelz A. G., Taylor Z. D., Wallace V. P., Zeitler J. A., Sibik J., Korter T. M., Ellison B., Rea S., Goldsmith P., Cooper K. B., Appleby R., Pardo D., Huggard P. G., Krozer V., Shams H., Fice M., Renaud C., Seeds A., Stohr A., Naftaly M., Ridler N. M., Clarke R., Cunningham J. E., Johnston M. B., “The 2017 terahertz science and technology roadmap,” J. Phys. D: Appl. Phys. 50(4), 043001 (2017). 10.1088/1361-6463/50/4/043001 [DOI] [Google Scholar]
- 3.Sizov F. F., “Infrared and terahertz in biomedicine,” Semicond. Phys., Quantum Electron. Optoelectron. 20(3), 273–283 (2017). 10.15407/spqeo20.03.273 [DOI] [Google Scholar]
- 4.Son J. H., Oh S. J., Cheon H., “Potential clinical applications of terahertz radiation,” J. Appl. Phys. 125(19), 190901 (2019). 10.1063/1.5080205 [DOI] [Google Scholar]
- 5.Tao Y. H., Fitzgerald A. J., Wallace V. P., “Non-contact, non-destructive testing in various industrial sectors with terahertz technology,” Sensors 20(3), 712 (2020). 10.3390/s20030712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaytsev K. I., Dolganova I. N., Chernomyrdin N. V., Katyba G. M., Gavdush A. A., Cherkasova O. P., Komandin G. A., Shchedrina M. A., Khodan A. N., Ponomarev D. S., Reshetov I. V., Karasik V. E., Skorobogatiy M., Kurlov V. N., Tuchin V. V., “The progress and perspectives of terahertz technology for diagnosis of neoplasms: a review,” J. Opt. 22(1), 013001 (2020). 10.1088/2040-8986/ab4dc3 [DOI] [Google Scholar]
- 7.Yang Y., Shutler A., Grischkowsky D., “Measurement of the transmission of the atmosphere from 0.2 to 2 THz,” Opt. Express 19(9), 8830–8838 (2011). 10.1364/OE.19.008830 [DOI] [PubMed] [Google Scholar]
- 8.Slocum D. M., Goyette T. M., Giles R. H., Nixon W. E., “Experimental determination of terahertz atmospheric absorption parameters,” Proc. SPIE 9483, 94830O (2015). 10.1117/12.2176455 [DOI] [Google Scholar]
- 9.Wilmink G. J., Grundt J. E., “Invited review article: current state of research on biological effects of terahertz radiation,” J. Infrared, Millimeter, Terahertz Waves 32(10), 1074–1122 (2011). 10.1007/s10762-011-9794-5 [DOI] [Google Scholar]
- 10.Hintzsche H., Stopper H., “Effects of terahertz radiation on biological systems,” Crit. Rev. Environ. Sci. Technol. 42(22), 2408–2434 (2012). 10.1080/10643389.2011.574206 [DOI] [Google Scholar]
- 11.Fedorov V. I., Serdyukov D. S., Cherkasova O. P., Popova S. S., Nemova E. F., “The influence of terahertz radiation on the cell's genetic apparatus,” J. Opt. Technol. 84(8), 509–514 (2017). 10.1364/JOT.84.000509 [DOI] [Google Scholar]
- 12.Il’ina I. V., Sitnikov D. S., Agranat M. B., “State-of-the-art of studies of the effect of terahertz radiation on living biological systems,” High Temp. 56(5), 789–810 (2018). 10.1134/S0018151X18050127 [DOI] [Google Scholar]
- 13.Cherkasova O. P., Serdyukov D. S., Ratushnyak A. S., Nemova E. F., Kozlov E. N., Shidlovsky Y. V., Zaitsev K. I., Tuchin V. V., “Mechanisms underlying the terahertz radiation effects on cells,” Opt. Spectrosc. 128(6), 855–866 (2020). 10.1134/S0030400X20060041 [DOI] [Google Scholar]
- 14.Williams R., Schofield A., Holder G., Downes J., Edgar D., Harrison P., Siggel-King M., Surman M., Dunning D., Hill S., Holder D., Jackson F., Jones J., McKenzie J., Saveliev Y., Thomsen N., Williams P., Weightman P., “The influence of high intensity terahertz radiation on mammalian cell adhesion, proliferation and differentiation,” Phys. Med. Biol. 58(2), 373–391 (2013). 10.1088/0031-9155/58/2/373 [DOI] [PubMed] [Google Scholar]
- 15.Koyama S., Narita E., Shimizu Y., Shiina T., Taki M., Shinohara N., Miyakoshi J., “Twenty four-hour exposure to a 0.12 THz electromagnetic field does not affect the genotoxicity, morphological changes, or expression of heat shock protein in HCE-T cells,” Int. J. Environ. Res. Public Health 13(8), 793 (2016). 10.3390/ijerph13080793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaekashiwa N., Yoshida H., Otsuki S., Hayashi S., Kawase K., “Verification of non-thermal effects of 0.3–0.6 THz-waves on human cultured cells,” Photonics 6(1), 33 (2019). 10.3390/photonics6010033 [DOI] [Google Scholar]
- 17.Hintzsche H., Jastrow C., Kleine-Ostmann T., Stopper H., Schmid E., Schrader T., “Terahertz radiation induces spindle disturbances in human hamster hybrid cells,” Radiat. Res. 175(5), 569–574 (2011). 10.1667/RR2406.1 [DOI] [PubMed] [Google Scholar]
- 18.Titova L. V., Ayesheshim A. K., Golubov A., Fogen D., Rodriguez Juarez R., Hegmann F. A., Kovalchuk O., “Intense THz pulses cause H2AX phosphorylation and activate DNA damage response in human skin tissue,” Biomed. Opt. Express 4(4), 559–568 (2013). 10.1364/BOE.4.000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franchini V., De Sanctis S., Marinaccio J., De Amicis A., Coluzzi E., Di Cristofaro S., Lista F., Regalbuto E., Doria A., Giovenale E., Gallerano G. P., Bei R., Benvenuto M., Masuelli L., Udroiu I., Sgura A., “Study of the effects of 0.15 terahertz radiation on genome integrity of adult fibroblasts,” Environ. Mol. Mutagen. 59(6), 476–487 (2018). 10.1002/em.22192 [DOI] [PubMed] [Google Scholar]
- 20.Alexandrov B. S., Phipps M. L., Alexandrov L. B., Booshehri L. G., Erat A., Zabolotny J., Mielke C. H., Chen H. T., Rodriguez G., Rasmussen KØ, Martinez J. S., Bishop A. R., Usheva A., “Specificity and heterogeneity of terahertz radiation effect on gene expression in mouse mesenchymal stem cells,” Sci. Rep. 3(1), 1184 (2013). 10.1038/srep01184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K. T., Park J., Jo S. J., Jung S., Kwon O. S., Gallerano G. P., Park W. Y., Park G. S., “High-power femtosecond-terahertz pulse induces a wound response in mouse skin,” Sci. Rep. 3(1), 2296 (2013). 10.1038/srep02296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Titova L. V., Ayesheshim A. K., Golubov A., Rodriguez-Juarez R., Woycicki R., Hegmann F. A., Kovalchuk O., “Intense THz pulses down-regulate genes associated with skin cancer and psoriasis: a new therapeutic avenue?” Sci. Rep. 3(1), 2363 (2013). 10.1038/srep02363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogomazova A. N., Vassina E. M., Goryachkovskaya T. N., Popik V. M., Sokolov A. S., Kolchanov N. A., Lagarkova M. A., Kiselev S. L., Peltek S. E., “No DNA damage response and negligible genome-wide transcriptional changes in human embryonic stem cells exposed to terahertz radiation,” Sci. Rep. 5(1), 7749 (2015). 10.1038/srep07749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Echchgadda I., Grundt J. E., Cerna C. Z., Roth C. C., Payne J. A., Ibey B. L., Wilmink G. J., “Terahertz radiation: a non-contact tool for the selective stimulation of biological responses in human cells,” IEEE Trans. Terahertz Sci. Technol. 6(1), 54–68 (2016). 10.1109/TTHZ.2015.2504782 [DOI] [Google Scholar]
- 25.Peltek S. E., Demidova E. V., Popik V. M., Goryachkovskaya T. N., “Stress-induced systems in Escherichia coli and their response to terahertz radiation,” Russ. J. Genet.: Appl. Res. 7(8), 858–868 (2017). 10.1134/S2079059717080019 [DOI] [Google Scholar]
- 26.Hough C. M., Purschke D. N., Huang C., Titova L. V., Kovalchuk O., Warkentin B. J., Hegmann F. A., “Global gene expression in human skin tissue induced by intense terahertz pulses,” Terahertz Sci. Technol. 11(1), 28–33 (2018). [Google Scholar]
- 27.Khlebodarova T. M., Tikunova N. V., Kachko A. V., Stepanenko I. L., Podkolodny N. L., Kolchanov N. A., “Application of bioinformatics resources for genosensor design,” J. Bioinf. Comput. Biol. 05(02b), 507–520 (2007). 10.1142/S0219720007002813 [DOI] [PubMed] [Google Scholar]
- 28.Merulla D., Hatzimanikatis V., van der Meer J. R., “Tunable reporter signal production in feedback-uncoupled arsenic bioreporters,” Microb. Biotechnol. 6(5), 503–514 (2013). 10.1111/1751-7915.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yagur-Kroll S., Schreuder E., Ingham C. J., Heideman R., Rosen R., Belkin S., “Miniature porous aluminum oxide-based flow-cell for online water quality monitoring using bacterial sensor cells,” Biosens. Bioelectron. 64, 625–632 (2015). 10.1016/j.bios.2014.09.076 [DOI] [PubMed] [Google Scholar]
- 30.Kim H. J., Lim J. W., Jeong H., Lee S. J., Lee D. W., Kim T., Lee S. J., “Development of a highly specific and sensitive cadmium and lead microbial biosensor using synthetic CadC-T7 genetic circuitry,” Biosens. Bioelectron. 79, 701–708 (2016). 10.1016/j.bios.2015.12.101 [DOI] [PubMed] [Google Scholar]
- 31.Demidova E. V., Goryachkovskaya T. N., Malup T. K., Bannikova S. V., Semenov A. I., Vinokurov N. A., Kolchanov N. A., Popik V. M., Peltek S. E., “Studying the non-thermal effects of terahertz radiation on E. coli/pKatG-GFP biosensor cells,” Bioelectromagnetics 34(1), 15–21 (2013). 10.1002/bem.21736 [DOI] [PubMed] [Google Scholar]
- 32.Demidova E. V., Goryachkovskaya T. N., Mescheryakova I. A., Malup T. K., Semenov A. I., Vinokurov N. A., Kolchanov N. A., Popik V. M., Peltek S. E., “Impact of terahertz radiation on stress sensitive genes of E. coli cell,” IEEE Trans. Terahertz Sci. Technol. 6(3), 435–441 (2016). 10.1109/TTHZ.2016.2532344 [DOI] [Google Scholar]
- 33.Lengeler J. W., Postma P. W., Biology of the Prokaryotes, Drews G., Lengeler J. W., Schlegel H. G., eds. (Thieme, 1999), pp. 491–523. [Google Scholar]
- 34.Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., 3rd, Smith H. O., “Enzymatic assembly of DNA molecules up to several hundred kilobases,” Nat. Methods 6(5), 343–345 (2009). 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 35.PTurboGFP-B vector (Evrogen), URL: https://evrogen.com/vector-descriptions/pTurboGFP-B/pTurboGFP-B.pdf. Accessed 01 June 2020.
- 36.PTurboYFP-B vector (Evrogen), URL: https://evrogen.com/vector-descriptions/pTurboYFP-B/pTurboYFP-B.pdf. Accessed 01 June 2020.
- 37.Green fluorescent protein TurboGFP (Evrogen), URL: https://evrogen.com/protein-descriptions/TurboGFP-description.pdf. Accessed 01 June 2020.
- 38.Yellow fluorescent protein TurboYFP (Evrogen), URL: https://evrogen.com/protein-descriptions/TurboYFP-description.pdf. Accessed 01 June 2020.
- 39.Shagin D. A., Barsova E. V., Yanushevich Y. G., Fradkov A. F., Lukyanov K. A., Labas Y. A., Semenova T. N., Ugalde J. A., Meyers A., Nunez J. M., Widder E. A., Lukyanov S. A., Matz M. V., “GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity,” Mol. Biol. Evol. 21(5), 841–850 (2004). 10.1093/molbev/msh079 [DOI] [PubMed] [Google Scholar]
- 40.Kulipanov G. N., Bagryanskaya E. G., Chesnokov E. N., Choporova Y. Y., Gerasimov V. V., Getmanov Y. V., Kiselev S. L., Knyazev B. A., Kubarev V. V., Peltek S. E., Popik V. M., Salikova T. V., Scheglov M. A., Seredniakov S. S., Shevchenko O. A., Skrinsky A. N., Veber S. L., Vinokurov N. A., “Novosibirsk free electron laser — facility description and recent experiments,” IEEE Trans. Terahertz Sci. Technol. 5(5), 798–809 (2015). 10.1109/TTHZ.2015.2453121 [DOI] [Google Scholar]
- 41.Kuryshev G. L., Kovchavizev A. P., Vainer B. G., Guzev A. A., Bazovkin V. M., Stroganov A. S., Subbotin I. M., Zakharov I. M., Efimov V. M., Postnikov K. O., Lee I. I., Valisheva N. A., Panova Z. V., “Medical infrared imaging system based on a 128×128 focal plane array for 2.8–3.05 µm spectral range,” Optoelectronics, Instrumentation and Data Processing 4, 5–10 (1998). [Google Scholar]
- 42.Terahertz sources (Terasense Group Inc.), URL: https://terasense.com/products/terahertz-sources. Accessed 01 June 2020.
- 43.Oshepkov D. Y., Goryachkovskaya T. N., Meshcheryakova I. A., Kiseleva E. V., Rozanov A. S., Bannikova S. V., Serdyukov D. S., Vasiliev G. V., Bryanskaya A. V., Vinokurov N. A., Popik V. M., Peltek S. E., “E. coli cells under impact of terahertz radiation,” NCBI Gene Expression Omnibus (GEO) database (2020), https://www.ncbi.nlm.nih.gov/bioproject/PRJNA648263.
- 44.Lehti T. A., Bauchart P., Dobrindt U., Korhonen T. K., Westerlund-Wikström B., “The fimbriae activator MatA switches off motility in Escherichia coli by repression of the flagellar master operon flhDC,” Microbiology (London, U. K.) 158(6), 1444–1455 (2012). 10.1099/mic.0.056499-0 [DOI] [PubMed] [Google Scholar]
- 45.Itou J., Eguchi Y., Utsumi R., “Molecular mechanism of transcriptional cascade initiated by the EvgS/EvgA system in Escherichia coli K-12,” Biosci., Biotechnol., Biochem. 73(4), 870–878 (2009). 10.1271/bbb.80795 [DOI] [PubMed] [Google Scholar]
- 46.Plumbridge J., Pellegrini O., “Expression of the chitobiose operon of Escherichia coli is regulated by three transcription factors: NagC, ChbR and CAP,” Mol. Microbiol. 52(2), 437–449 (2004). 10.1111/j.1365-2958.2004.03986.x [DOI] [PubMed] [Google Scholar]
- 47.Smolyanskaya O. A., Chernomyrdin N. V., Konovko A. A., Zaytsev K. I., Ozheredov I. A., Cherkasova O. P., Nazarov M. M., Guillet J.-P., Kozlov S. A., Kistenev Y. V., Coutaz J.-L., Mounaix P., Vaks V. L., Son J. H., Cheon H., Wallace V. P., Feldman Y., Popov I., Yaroslavsky A. N., Shkurinov A. P., Tuchin V. V., “Terahertz biophotonics as a tool for studies of dielectric and spectral properties of biological tissues and liquids,” Prog. Quantum Electron. 62, 1–77 (2018). 10.1016/j.pquantelec.2018.10.001 [DOI] [Google Scholar]
- 48.Xu J., Plaxco K. W., Allen S. J., “Absorption spectra of liquid water and aqueous buffers between 0.3 and 3.72 THz,” J. Chem. Phys. 124(3), 036101 (2006). 10.1063/1.2151267 [DOI] [PubMed] [Google Scholar]
- 49.Hough C. M., Purschke D. N., Huang C., Titova L. V., Kovalchuk O., Warkentin B. J., Hegmann F. A., “Intensity-dependent suppression of calcium signaling in human skin tissue models induced by intense THz pulses,” in 43rd International Conference on Infrared, Millimeter, and Terahertz Waves (IRMMW-THz) (2018), pp. 218–221. [Google Scholar]
- 50.Echchgadda I., Cernab C. Z., Sloanb M. A., Elamc D. P., Ibeya B. L., “Effects of different terahertz frequencies on gene expression in human keratinocytes,” Proc. SPIE 9321, 93210Q (2015). 10.1117/12.2082542 [DOI] [Google Scholar]
- 51.Alexandrov B. S., Rasmussen K. O., Bishop A. R., Usheva A., Alexandrov L. B., Chong S., Dagon Y., Booshehri L. G., Mielke C. H., Phipps M. L., Martinez J. S., Chen H. T., Rodriguez G., “Non-thermal effects of terahertz radiation on gene expression in mouse stem cells,” Biomed. Opt. Express 2(9), 2679–2689 (2011). 10.1364/BOE.2.002679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehti T. A., Bauchart P., Heikkinen J., Hacker J., Korhonen T. K., Dobrindt U., Westerlund-Wikström B., “Mat fimbriae promote biofilm formation by meningitis-associated Escherichia coli,” Microbiology (London, U. K.) 156(8), 2408–2417 (2010). 10.1099/mic.0.039610-0 [DOI] [PubMed] [Google Scholar]
- 53.Soutourina O. A., Bertin P. N., “Regulation cascade of flagellar expression in Gram-negative bacteria,” FEMS Microbiol. Rev. 27(4), 505–523 (2003). 10.1016/S0168-6445(03)00064-0 [DOI] [PubMed] [Google Scholar]
- 54.Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J., “Bacterial biofilms in nature and disease,” Annu. Rev. Microbiol. 41(1), 435–464 (1987). 10.1146/annurev.mi.41.100187.002251 [DOI] [PubMed] [Google Scholar]
- 55.Schembri M. A., Kjaergaard K., Klemm P., “Global gene expression in Escherichia coli biofilms,” Mol. Microbiol. 48(1), 253–267 (2003). 10.1046/j.1365-2958.2003.03432.x [DOI] [PubMed] [Google Scholar]
- 56.Kachroo A. H., Kancherla A. K., Singh N. S., Varshney U., Mahadevan S., “Mutations that alter the regulation of the chb operon of Escherichia coli allow utilization of cellobiose,” Mol. Microbiol. 66(6), 1382–1395 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Sezonov G., Joseleau-Petit D., D’Ari R., “Escherichia coli physiology in Luria-Bertani broth,” J. Bacteriol. 189(23), 8746–8749 (2007). 10.1128/JB.01368-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hua Q., Yang C., Oshima T., Mori H., Shimizu K., “Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures,” Appl. Environ. Microbiol. 70(4), 2354–2366 (2004). 10.1128/AEM.70.4.2354-2366.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burton N. A., Johnson M. D., Antczak P., Robinson A., Lund P. A., “Novel aspects of the acid response network of E. coli K-12 are revealed by a study of transcriptional dynamics,” J. Mol. Biol. 401(5), 726–742 (2010). 10.1016/j.jmb.2010.06.054 [DOI] [PubMed] [Google Scholar]
- 60.Ma Z., Masuda N., Foster J. W., “Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli,” J. Bacteriol. 186(21), 7378–7389 (2004). 10.1128/JB.186.21.7378-7389.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamanaka Y., Oshima T., Ishihama A., Yamamoto K., “Characterization of the YdeO regulon in Escherichia coli,” PLoS One 9(11), e111962 (2014). 10.1371/journal.pone.0111962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foster J. W., “Escherichia coli acid resistance: tales of an amateur acidophile,” Nat. Rev. Microbiol. 2(11), 898–907 (2004). 10.1038/nrmicro1021 [DOI] [PubMed] [Google Scholar]
- 63.Masuda N., Church G. M., “Regulatory network of acid resistance genes in Escherichia coli,” Mol. Microbiol. 48(3), 699–712 (2003). 10.1046/j.1365-2958.2003.03477.x [DOI] [PubMed] [Google Scholar]
- 64.Menon N. K., Robbins J., Peck Jr H. D., Chatelus C. Y., Choi E. S., Przybyla A. E., “Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames,” J. Bacteriol. 172(4), 1969–1977 (1990). 10.1128/JB.172.4.1969-1977.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dassa J., Fsihi H., Marck C., Dion M., Kieffer-Bontemps M., Boquet P. L., “A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA),” Mol. Gen. Genet. 229(3), 341–352 (1991). 10.1007/BF00267454 [DOI] [PubMed] [Google Scholar]
- 66.Rahav-Manor O., Carmel O., Karpel R., Taglicht D., Glaser G., Schuldiner S., Padan E., “NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaA, the sodium proton antiporter gene in Escherichia coli,” J. Biol. Chem. 267(15), 10433–10438 (1992). [PubMed] [Google Scholar]
- 67.Gustavsson N., Diez A., Nyström T., “The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage,” Mol. Microbiol. 43(1), 107–117 (2002). 10.1046/j.1365-2958.2002.02720.x [DOI] [PubMed] [Google Scholar]
- 68.Nachin L., Nannmark U., Nyström T., “Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility,” J. Bacteriol. 187(18), 6265–6272 (2005). 10.1128/JB.187.18.6265-6272.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.