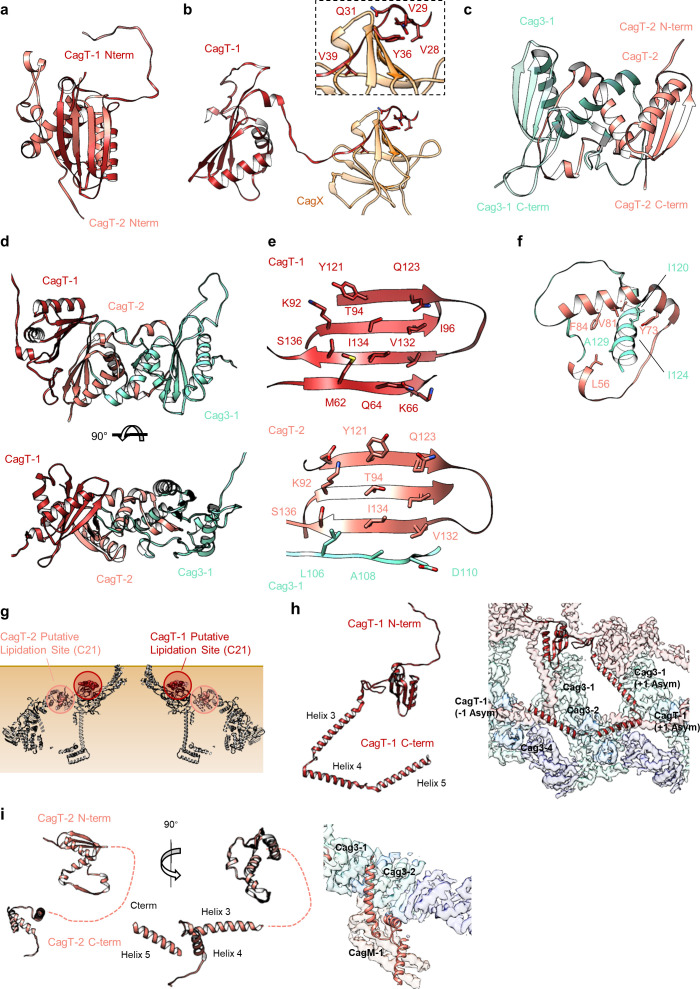
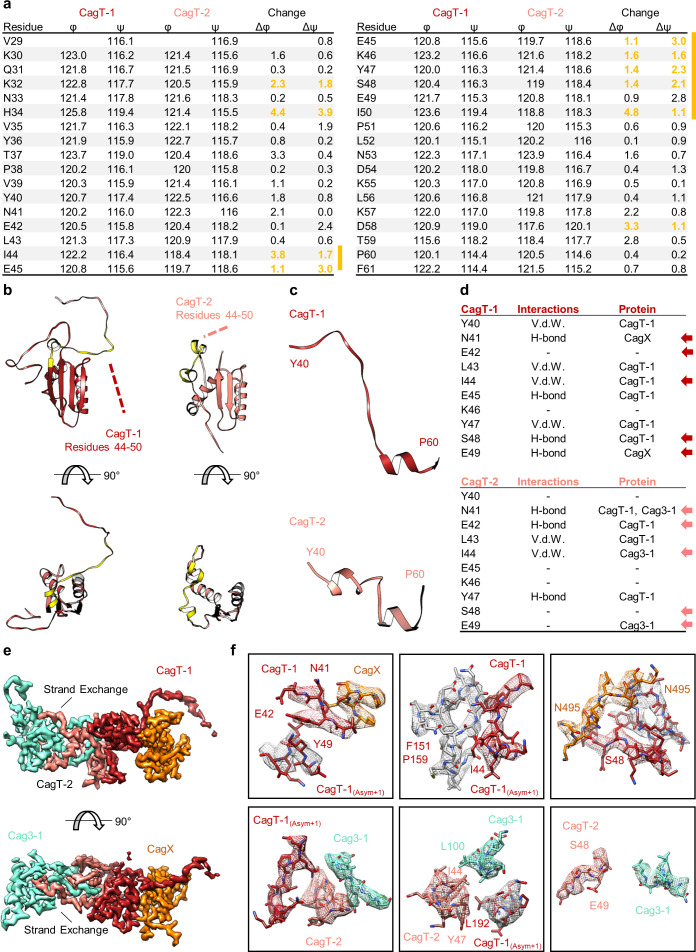
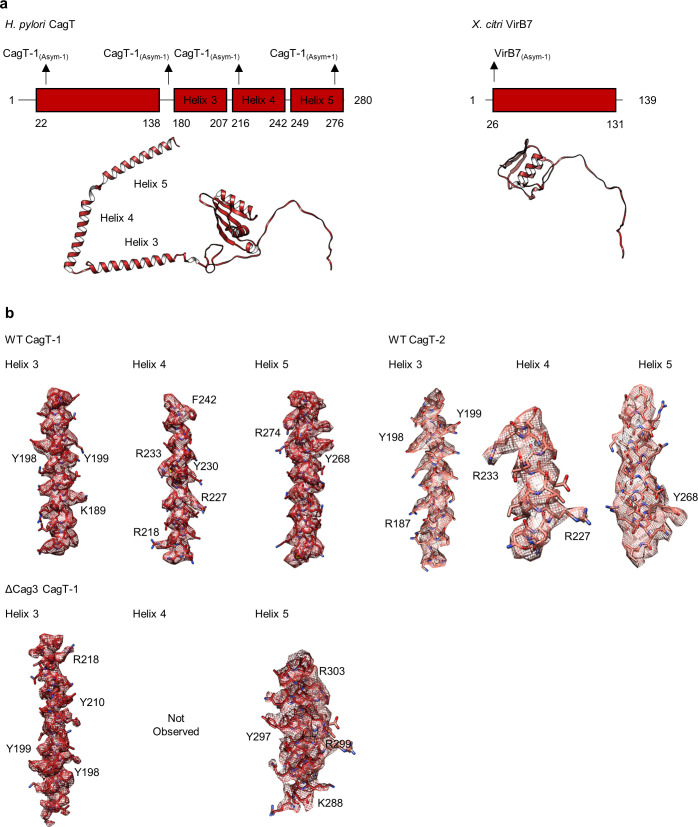
Figure 4. The N- and C-termini from CagT-1 and CagT-2 are positioned differently.
(a–c) The N-terminus of CagT-1 extends inward toward the center of the map and interacts with CagX from the next asymmetric unit. The residues that contribute to this interaction are largely hydrophobic, as indicated in the inset panel. CagT-2 differs from CagT-1 in that the N-terminus of the protein extends outward toward the periphery of the map, forming the last strand of a β-sheet with Cag3-1. (d) The three proteins (CagT-1, CagT-2 and Cag3-1) have an interwoven architecture. (e) The interface that is formed between the three proteins consists of two β-sheets that include strands from all three molecules. (f) The interface of CagT-2 and Cag3-1 is a pair of α-helices that bury hydrophobic residues within the interface. (g) The position of the N-terminal loops of CagT-1 (red) and CagT-2 (salmon) are such that the putative lipidation sites are near the outer membrane. (h) The C-terminal α-helices of CagT-1 adopt an extended conformation (left) and interact with Cag3-1, Cag3-2, and Cag3-4 within the same asymmetric unit and Cag3-1 and CagT-1 in neighboring asymmetric units (right). (i) The C-terminal α-helices of CagT-2 are connected by an apparently flexible linker (left) and interact with Cag3-1, Cag3-2, and CagM-1 (right).