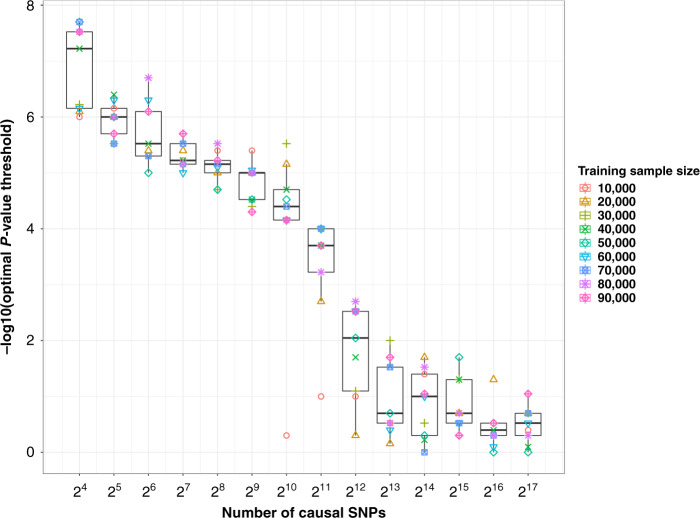
Fig. 3. The relationship between optimal GRS P-value threshold and number of causal SNPs.
Causal SNPs were selected from 1,037,804 HapMap3 SNPs. For each scenario, we generated a phenotype of 100,000 individuals based on a specified number of causal SNPs (e.g., 128) with heritability 0.09. We randomly selected 10,000 individuals as the test set. Based on the unselected individuals, we randomly chose 10,000, 20,000, 30,000, 40,000, 50,000, 60,000, 70,000, 80,000 and 90,000 individuals separately as training sets and used them to perform GWAS. We examined the performance of genetic risk score (based on LD clumping with 80 separate P-value thresholds) on the test set (Ntest = 10,000) and selected the optimal P-value threshold. Box plot shows the median (centre line), the interquartile range (box) and whiskers (±1.5 times interquartile range).