Figure 2. Comparison of structures of phosphorylated mouse and human IRF-3 bound to CBP.
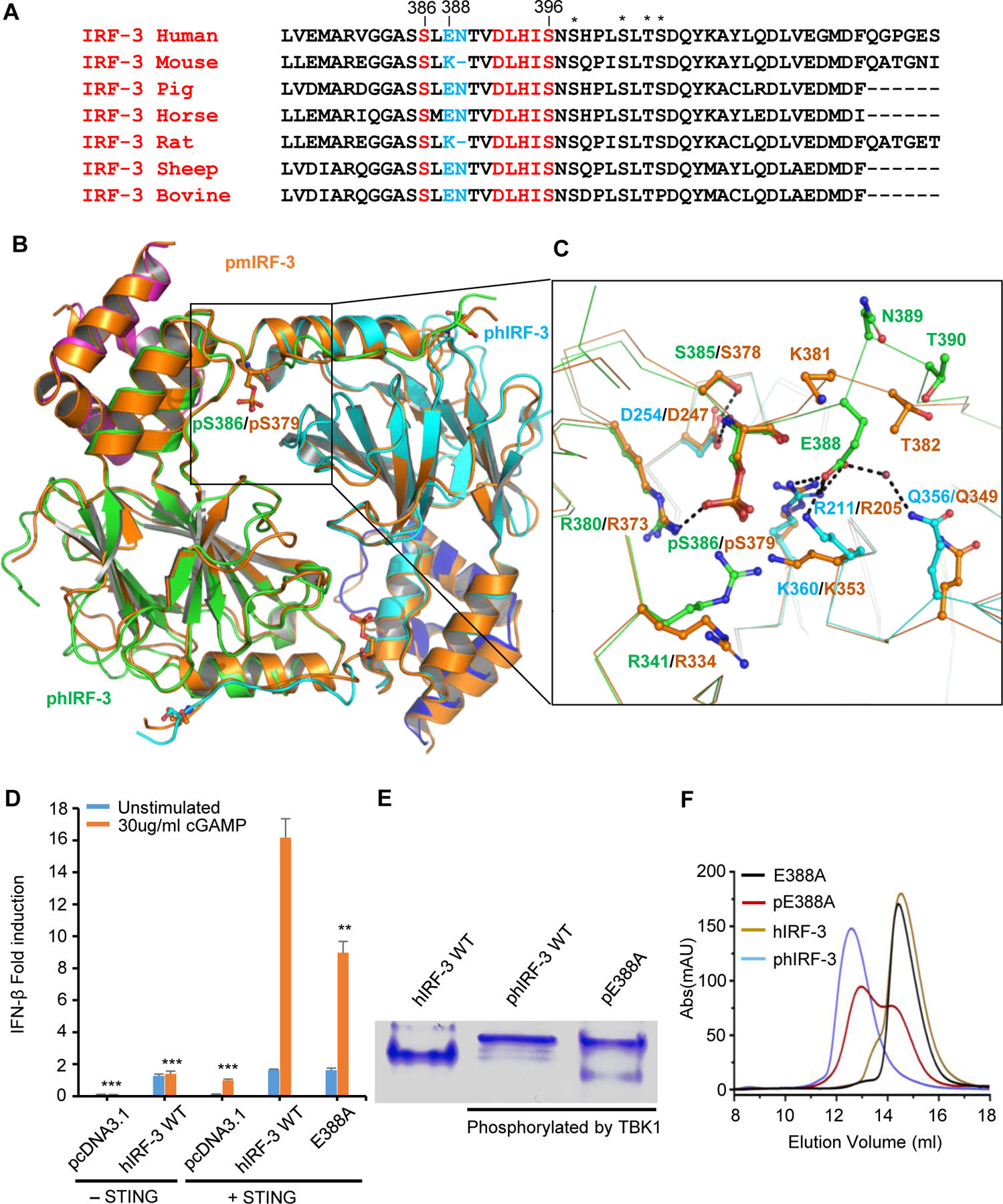
(A) Sequence alignment of C-terminal tail of IRF-3 across different species showing the conserved phosphorylation sites and the pLxIS motif in red. Residues corresponding to Glu388 and Asn389 of human IRF-3 are highlighted in blue. Other potential phosphorylation sites are indicated by the asterisks. (B) Comparison of structures of phosphorylated mouse and human IRF-3/CBP complexes. Phosphorylated mouse IRF-3/CBP is shown by the orange ribbon. Human IRF-3 dimer is colored in green and cyan with CBP bound to hIRF-3 in magenta and blue. Phosphorylated Ser379 of mIRF-3 and Ser386 of hIRF-3 are shown by the ball-and-stick models. (C) Distinct interactions between phosphorylated mouse and human IRF-3. Key residues mediating human and mouse IRF-3 dimerization are colored in green and orange, respectively. Residues interacting with Glu388 of hIRF-3 are in cyan. (D) IFN-β luciferase reporter assays showing the effect of E388A mutation on the signaling mediated by hIRF-3. The data are mean ± s.e.m. and representative of three independent assays. **P < 0.01, ***P < 0.001 values were calculated by comparisons of signals in cells transfected with E388A mutant and those transfected with wild-type IRF-3. (E) Native gel electrophoresis showing the dimerization of wild-type human IRF-3 and its E388A mutant upon phosphorylation. (F) Size-exclusion chromatography showing the effect of E388A mutation on the dimerization of hIRF-3 upon phosphorylation as compared to wild-type IRF-3.
