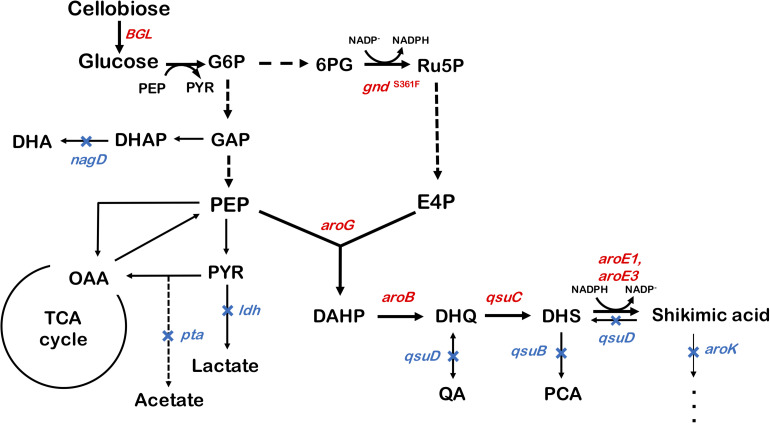
FIGURE 1.
Metabolic engineering of Corynebacterium glutamicum for shikimic acid production. The blue X indicates deletion of the nagD, qsuD, qsuB, qsuD, aroK, and pta genes. Genes involving shikimic acid synthesis pathways are indicated in red. G6P, glucose-6-phosphate; GAP, glyceraldehyde-3-phosphate; DHAP, 1,3-dihydroxyacetone phosphate; DHA, 1,3-dihydroxyacetone; PEP, phosphoenolpyruvate; PYR, pyruvate; OAA, oxaloacetate; Ru5P, ribulose-5-phosphate; E4P, erythrose 4-phosphate; DAHP, 3-deoxy-D-arabinoheptulosonate-7-phosphate; DHQ, 3-de-hydroquinate; DHS, 3-dehydroshikimate; PCA, protocatechuate; aroG, DAHP synthase; aroB, DHQ synthase; aroD, DHQ dehydratase; aroE1, aroE3, shikimate dehydrogenase; aroK, shikimate kinase; qsuB, DHS dehydratase; qsuD, QA/shikimate dehydrogenase; gnd, 6-phosphogluconate dehydrogenase; BGL, beta-glucosidase from Thermobifida fusca YX.