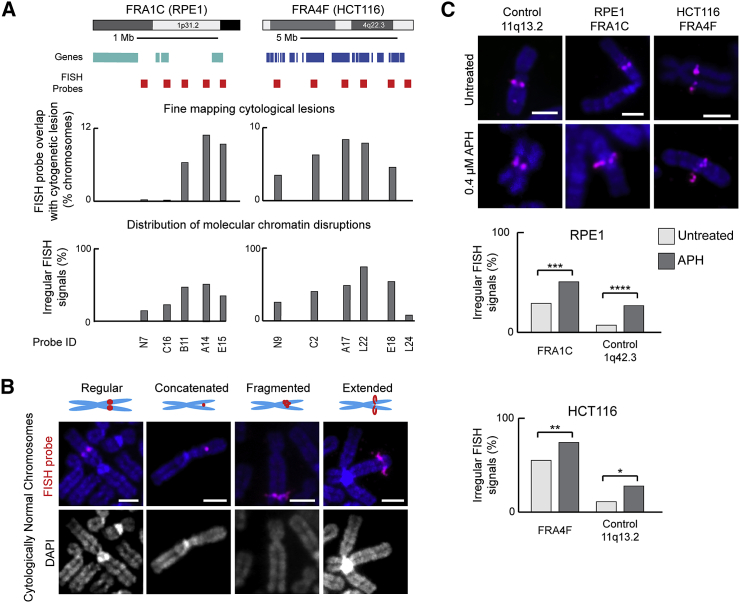
Figure 2.
Irregular Chromatin Structures at CFSs in the Presence and Absence of APH-Induced Replication Stress
(A) Quantification of FISH probe signals across FRA4F (HCT116 cells, n = 439) and FRA1C (RPE1 cells, n = 180) to finely map cytological lesions (top) and distribution of molecular chromatin disruptions (bottom). FISH probe IDs are described in the Method Details.
(B) Irregular FISH probe phenotypes (magenta) on cytogenetically normal chromosomes. Regular, symmetrical signals; concatenated, a single-signal sitting between the two sister chromatids; fragmented, multiple, asymmetric signals; extended, a signal extending beyond the DAPI-stained chromosome area. Scale bar, 2.5 μm.
(C) Chromosomes from untreated cells (top) or cells treated with APH (bottom), to induce replication stress, hybridized to FISH probes for a non-fragile locus 11q13.2 (probe ID: P21, n = 84 for HCT116, n = 87 for RPE1) or fragile-loci FRA4F (probe ID: A17; HCT116 cells, n = 123) and FRA1C (probe ID: A14; RPE1 cells, n = 146). Scale bar, 2.5 μm. Bottom, quantification of irregular FISH signals in the presence and absence of APH. p values for a χ2 test.
∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.