Figure 4.
Molecular Chromatin Disruptions at CFSs Are Not Remodeled for Mitosis
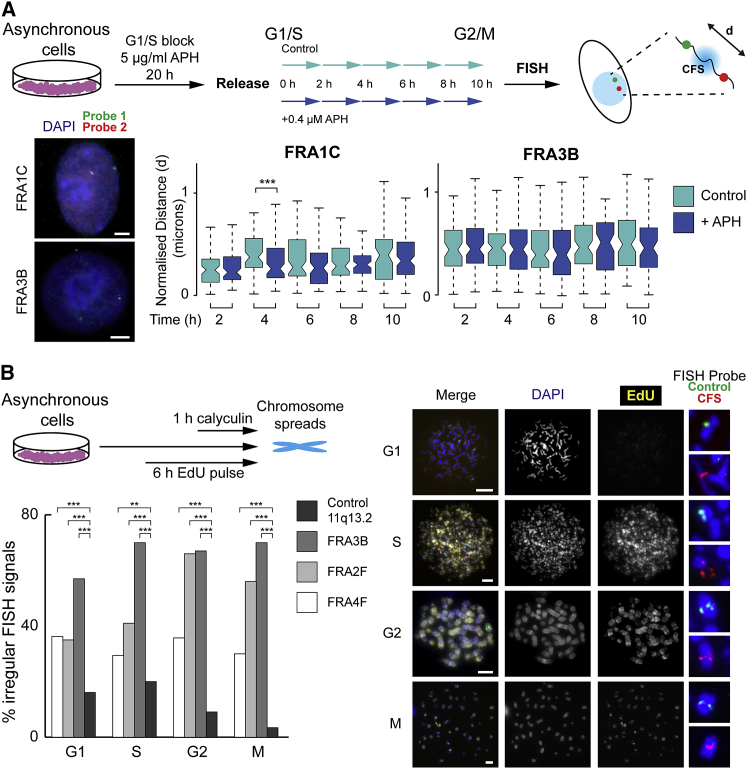
(A) Top, model detailing experimental strategy to analyze interphase chromatin folding at CFS regions. Cells progressing synchronously through the cell cycle were harvested every 2 h and fosmid probes approximately 1.5 Mb apart surrounding FRA1C (probes B3 and C1) or FRA3B (probes E4 and F5) were hybridized to nuclei and imaged. Bottom left, representative FISH images. Bottom right, boxplot of normalized inter-fosmid distances (d in drawing) between pairs of probes hybridized to nuclei (n > 60 nuclei for each time point). p values are for a Wilcoxon test.
(B) Depiction of premature chromosome condensation (PCC) assay (see Method Details) in HCT116 cells. Cells labeled with EdU (6 h) were condensed with calyculin (1 h), harvested, and hybridized to FISH probes for a control locus (11q13.2, probe P21) and CFSs (FRA3B, probe C2; FRA4F, probe A17; and FRA2F, probe K5). Right, representative chromosome images. Bottom, left, quantification of irregular FISH probe signals at FRA3B (n = 122 chromosomes), FRA2F (n = 136), FRA4F (n = 119), and 11q13.2 control probe (n = 116); p values for a χ2 test.
Scale bars, 2.5 μm. NS, not significant; ∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.