FIGURE 1.
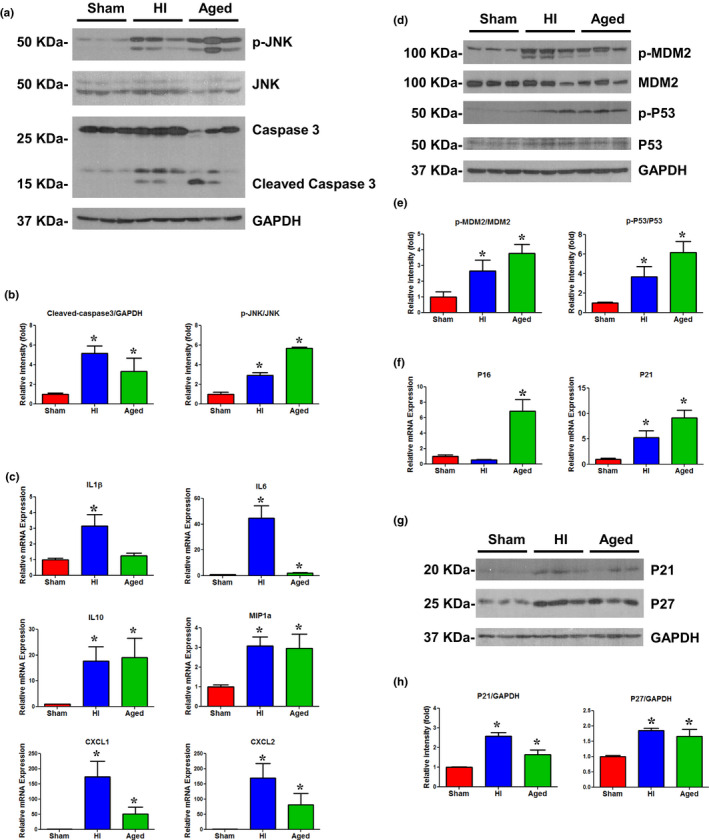
(a) Representative immunoblotting of p‐JNK, JNK, and Caspase3 in liver tissues of sham, HI, and aged rats. Loading control: GAPDH. (b) p‐JNK, JNK, cleaved‐Caspase3, and GAPDH band intensities (see Figure 1a) were quantified using Image J software (NIH). n = 6 animals/group, 2–3 technical replicates. *p < 0.05 vs. sham. (c) SASP gene expression was measured by real‐time PCR in the liver tissues of sham, HI, and aged rats. n = 5–6/group; two technical replicates; bars: mean ± SEM; *p < 0.05 vs. sham. (d) Representative immunoblotting of p‐MDM2, MDM2, p‐P53, and P53 in liver tissues of sham, HI, and aged rats. Loading control: GAPDH. (e) p‐MDM2, MDM2, p‐P53, P53, and GAPDH intensities (see Figure 1d) were quantified using Image J software (NIH). n = 6 animals/group, 2–3 technical replicates. *p < 0.05 vs. sham. (f) p16 and p21 gene expressions measured by real‐time PCR in the liver tissues of sham, HI, and aged rats. n = 5–6/group; two technical replicates; bars: mean ± SEM; *p < 0.05 vs. sham. (g) Representative immunoblotting of p21 and p27 in liver tissues of sham, HI, and aged rats. Loading control: GAPDH. (h) P21, p27, and GAPDH intensities (see Figure 1g) were quantified using Image J software (NIH). n = 6 animals/group, 2–3 technical replicates. *p < 0.05 vs. sham
