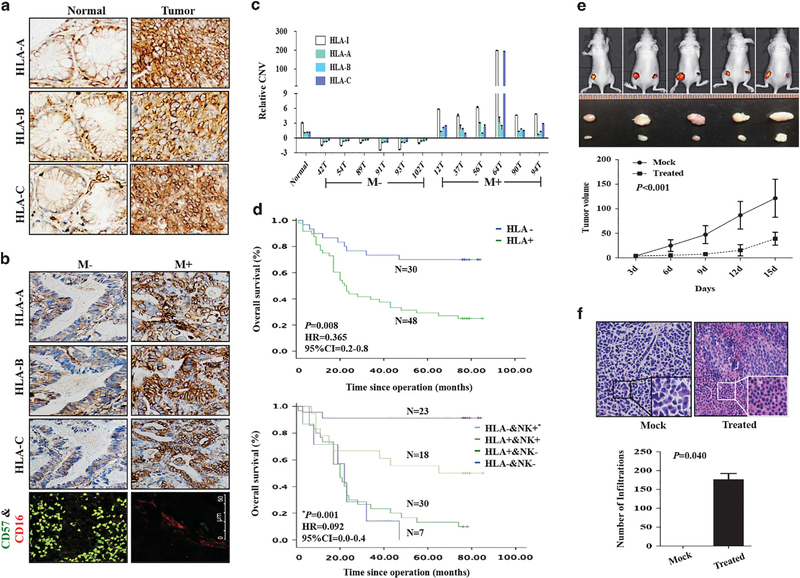
Figure 5.
Expression patterns of HLA-I in GC-matched tissues. (a) Immunohistochemistry of HLA-I in GC-matched tissues. (b) Immunohistochemistry of HLA-I and NK cells in GC tissues with or without metastasis. (c) qPCR detected relative CNV of HLA-A, -B and -C in GC-matched tissues from patients with or without metastasis. CNVs in tumors was normalized by matched adjacent normal tissues. The value of HLA-I is the sum value of HLA-A, -B and -C. (d, upper) Survival analysis of HLA-I. P-value, hazard ratio (HR) and 95% confidence interval (CI) were obtained from the multivariate analysis (Supplementary Table S3). (Lower) Survival analysis of HLA-I combined with NK cells. P-value, HR and 95% CI were obtained from the multivariate analysis (Supplementary Table S4). (e, upper) Nude mice of subcutaneous injection with BGC823 treated with IL-12 and antibody against HLA-I (right) or IgG (left). (Lower) Tumor growth curves. (f, upper left) HE staining of xenograft tumor tissues from mock group. Inner, the enlargement of tumor cells; upper right, HE staining of xenograft tumor tissues from treated group. Inner, the enlargement of NK cells around the tumor cells; (f, lower), the number of NK cells around the tumor cells in mock and treated groups.