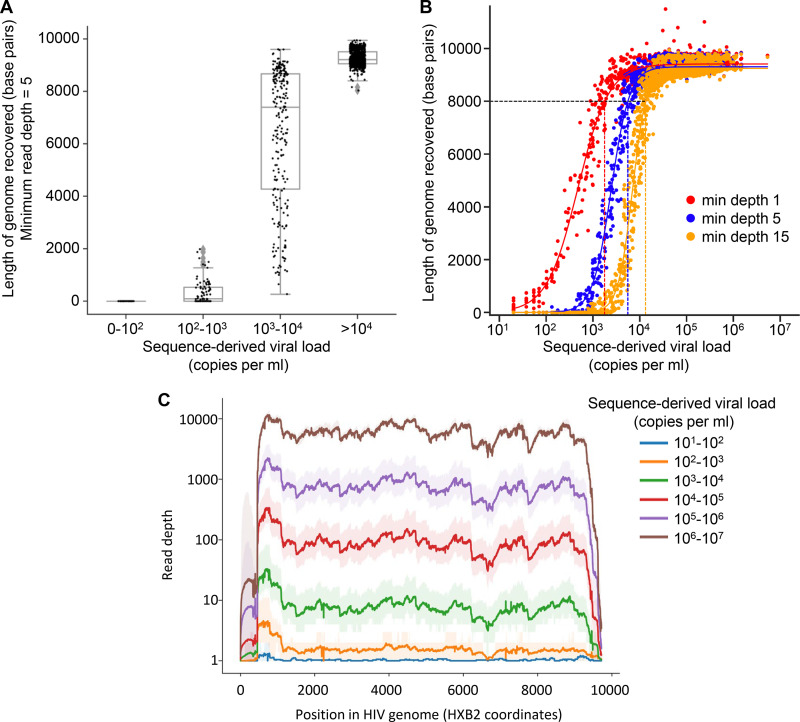
FIG 4.
Sequencing success is influenced by viral load. (A) The length of the HIV genomes reconstructed by shiver software, from paired-end Illumina reads, stratified by log viral load, showed reproducible whole-genome coverage for samples with sequence inferred viral loads of >4 log10 copies/ml and near-complete coverage for the majority of samples with VL between 3 and 4 log10 copies/ml. (B) The viral loads at which genome coverage exceed 8 kb with minimum depth thresholds of 1 read, 5 reads, and 15 reads (after removal of PCR duplicates) are shown by the intercepts of curves fitted using a sigmoid function. (C) The median (thick lines) and 95th percentile range (ribbons) of read-depth observed across the genome are shown for all samples, grouped by sequence-derived viral load.