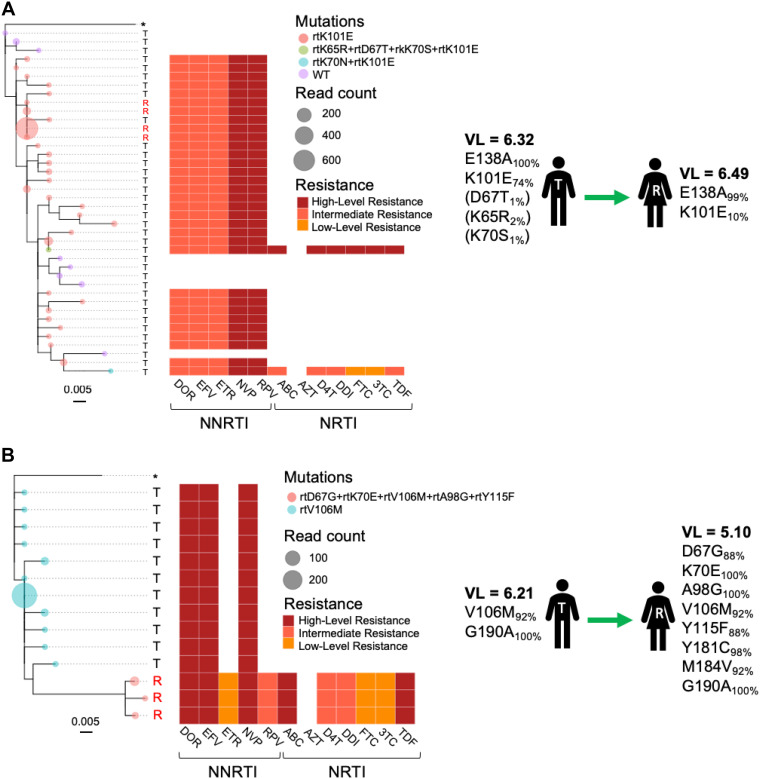
FIG 5.
Within-host phylogenetic trees of Illumina reads spanning drug resistance sites in pol. Phyloscanner software performs ancestral state reconstructions of phylogenetic trees generated from Illumina reads in “windows” across the genome in order to identify pairs consisting of transmitters (T) and recipients (R). Phylogenetic trees of reads spanning drug resistance mutations sites in pol are shown for two inferred transmission pairs (A and B). Tree tips (circles) are colored by the combinations of drug resistance mutations observed for each unique taxon and scaled to total read counts within each taxa (after removal of PCR duplicates). Heatmaps report the predicted drug susceptibilities for each read using the Stanford HIVdb classification. Sequence-derived viral loads (log 10 RNA copies/ml) and the complete list of resistance mutations with associated frequencies, observed across entire genomes, are shown for each individual. Mutations observed at frequencies below 5% are shown in parentheses.