Figure 1 -. Assessment of acute and chronic effects of dietary MR on EE in Control mice (Klbfl/fl), mice with adipocyte-specific Klb knockout (Klbfl/(Adip)), or mice with brain-specific Klb knockout (Klbfl/(CamK2a)).
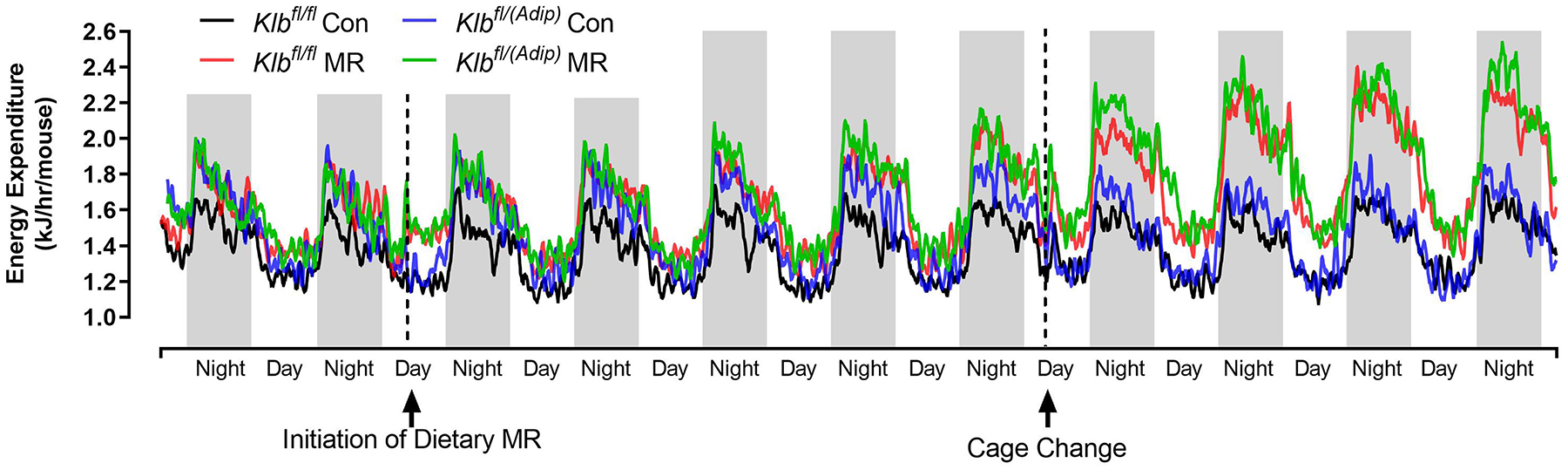
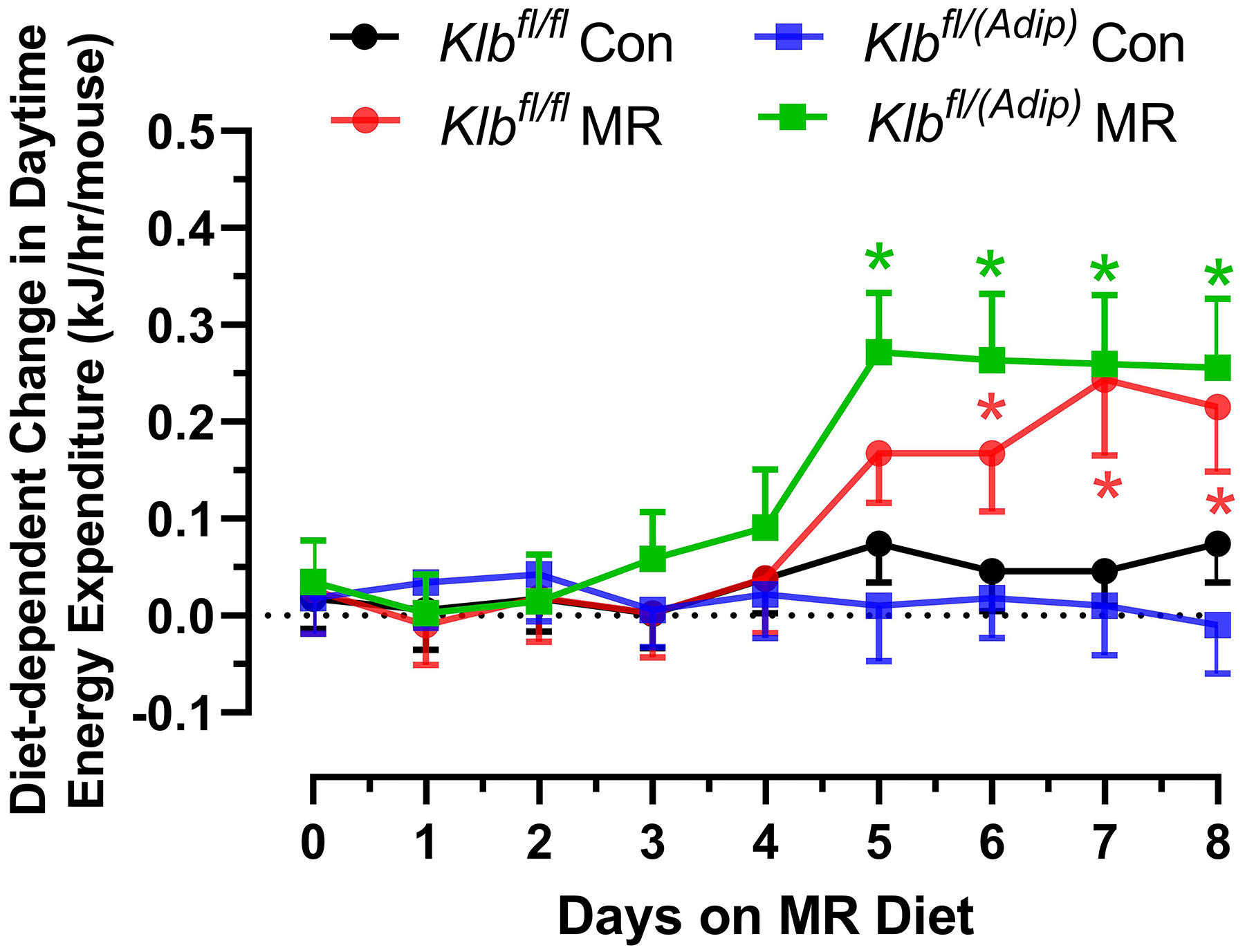
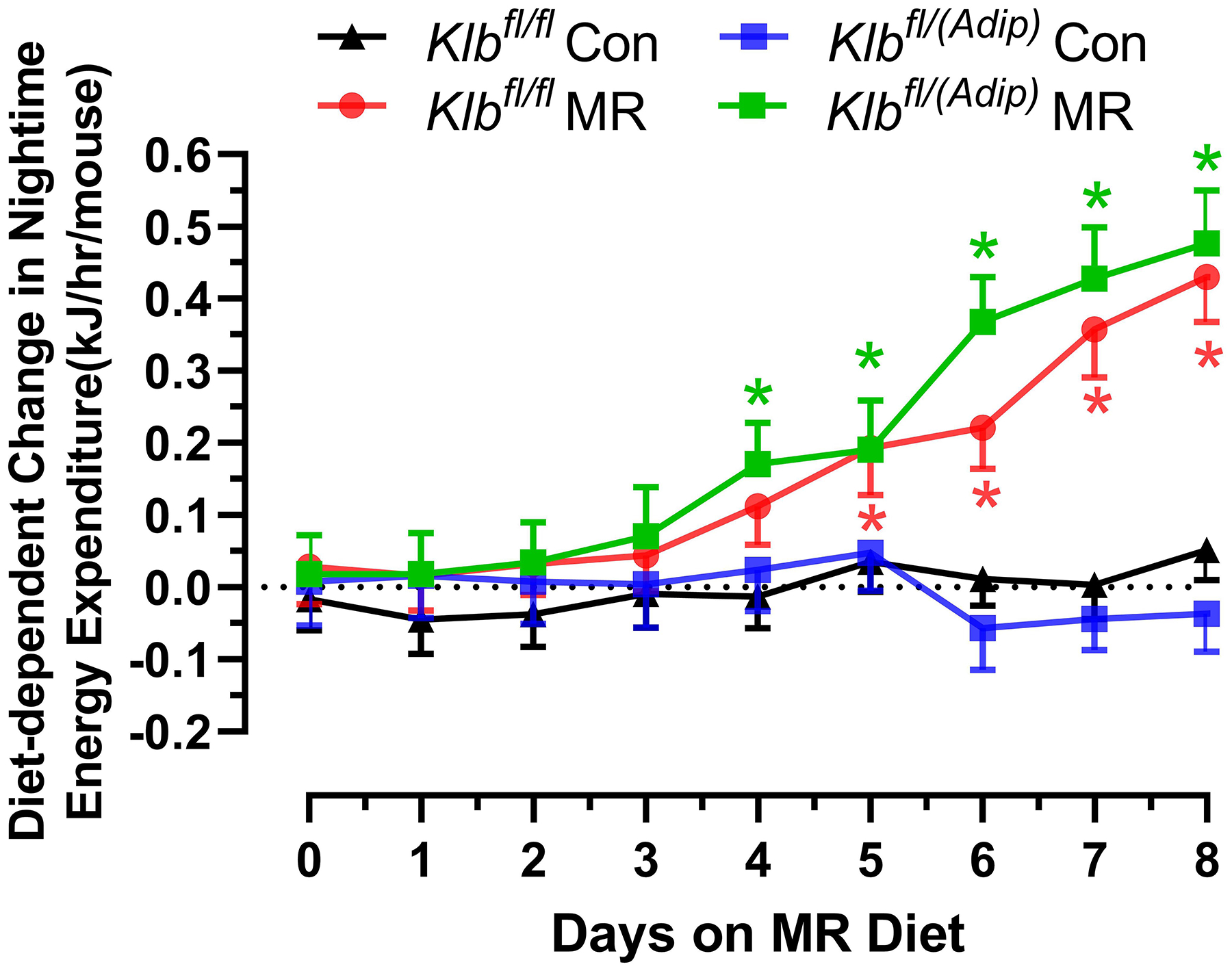
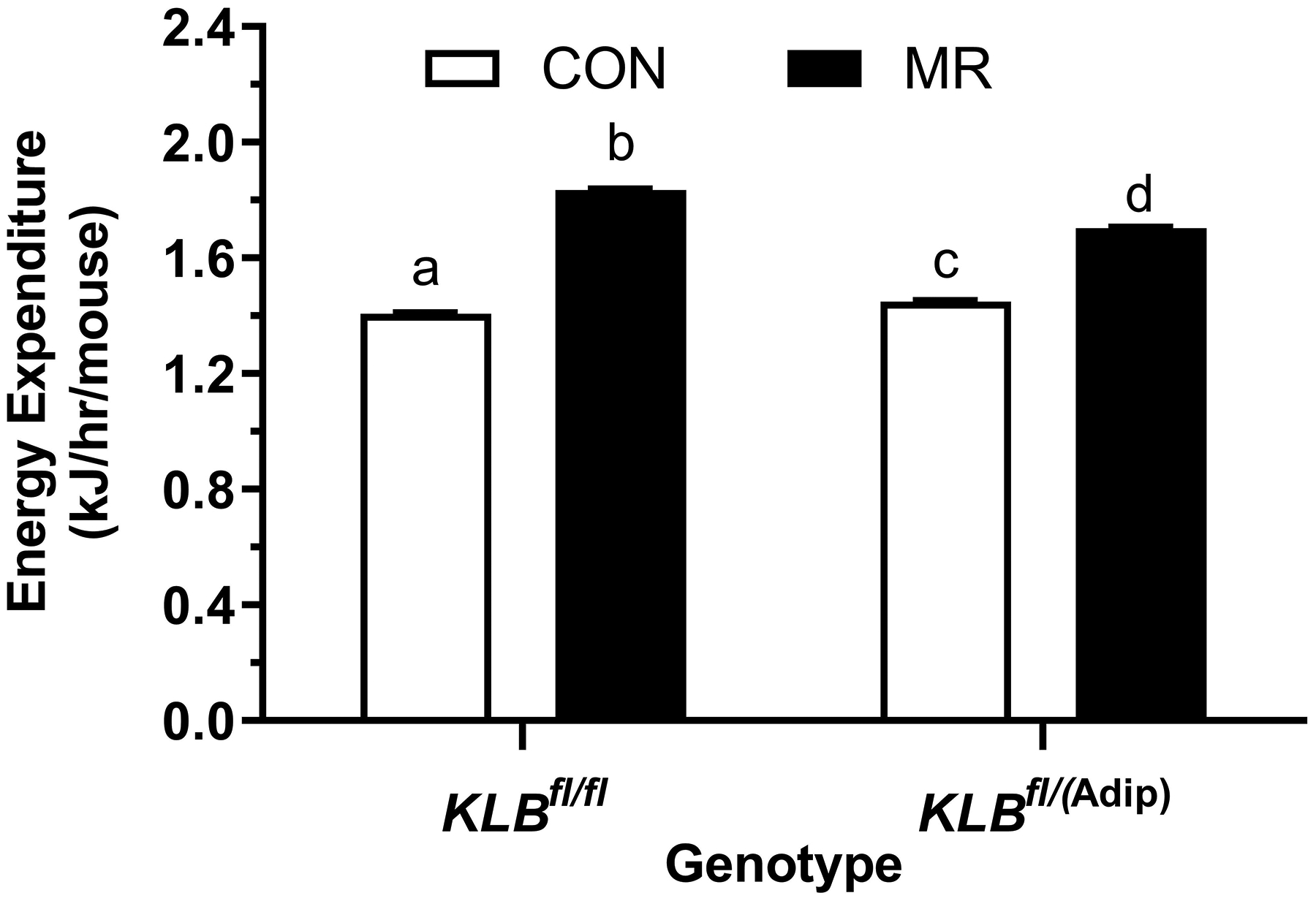
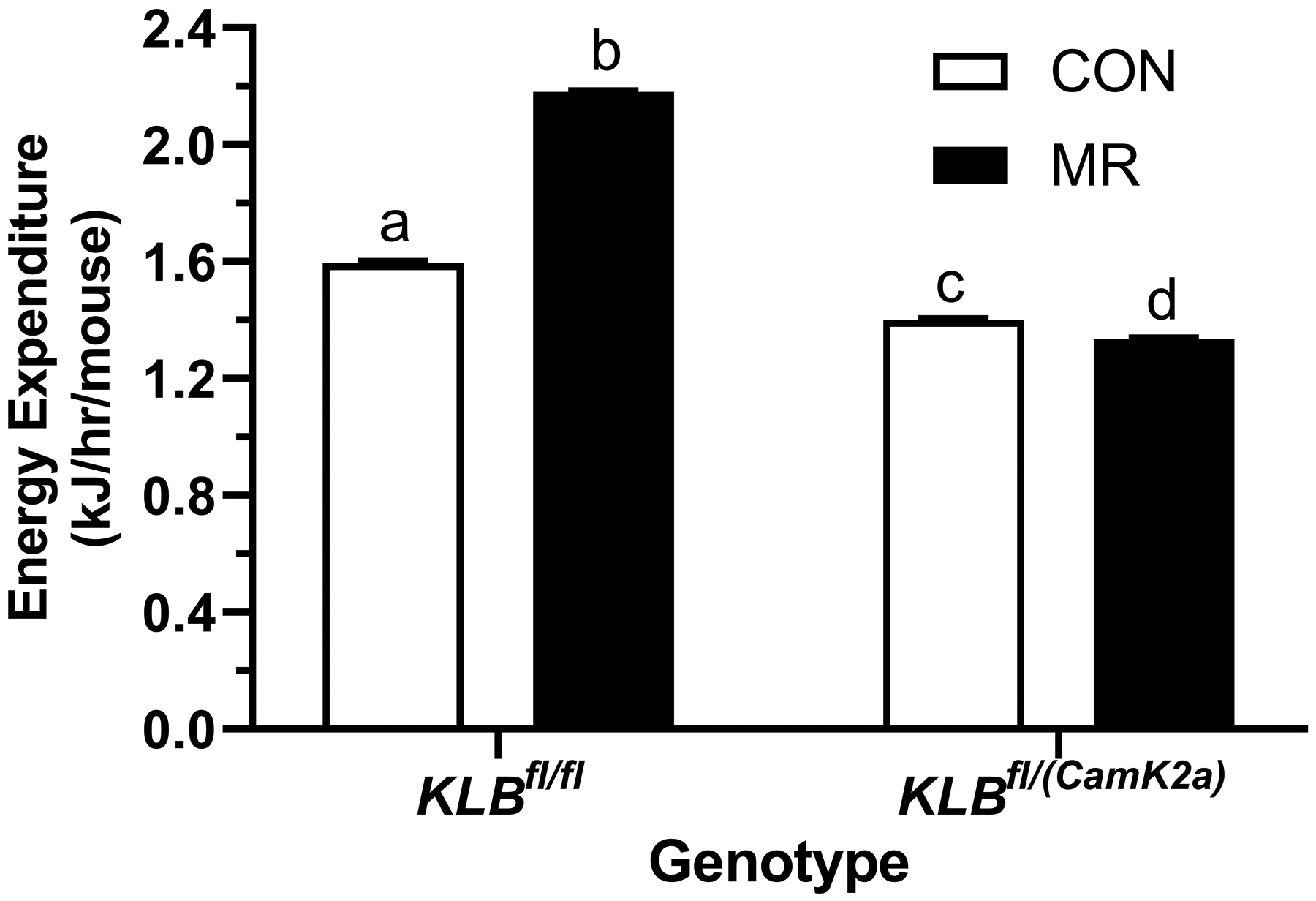
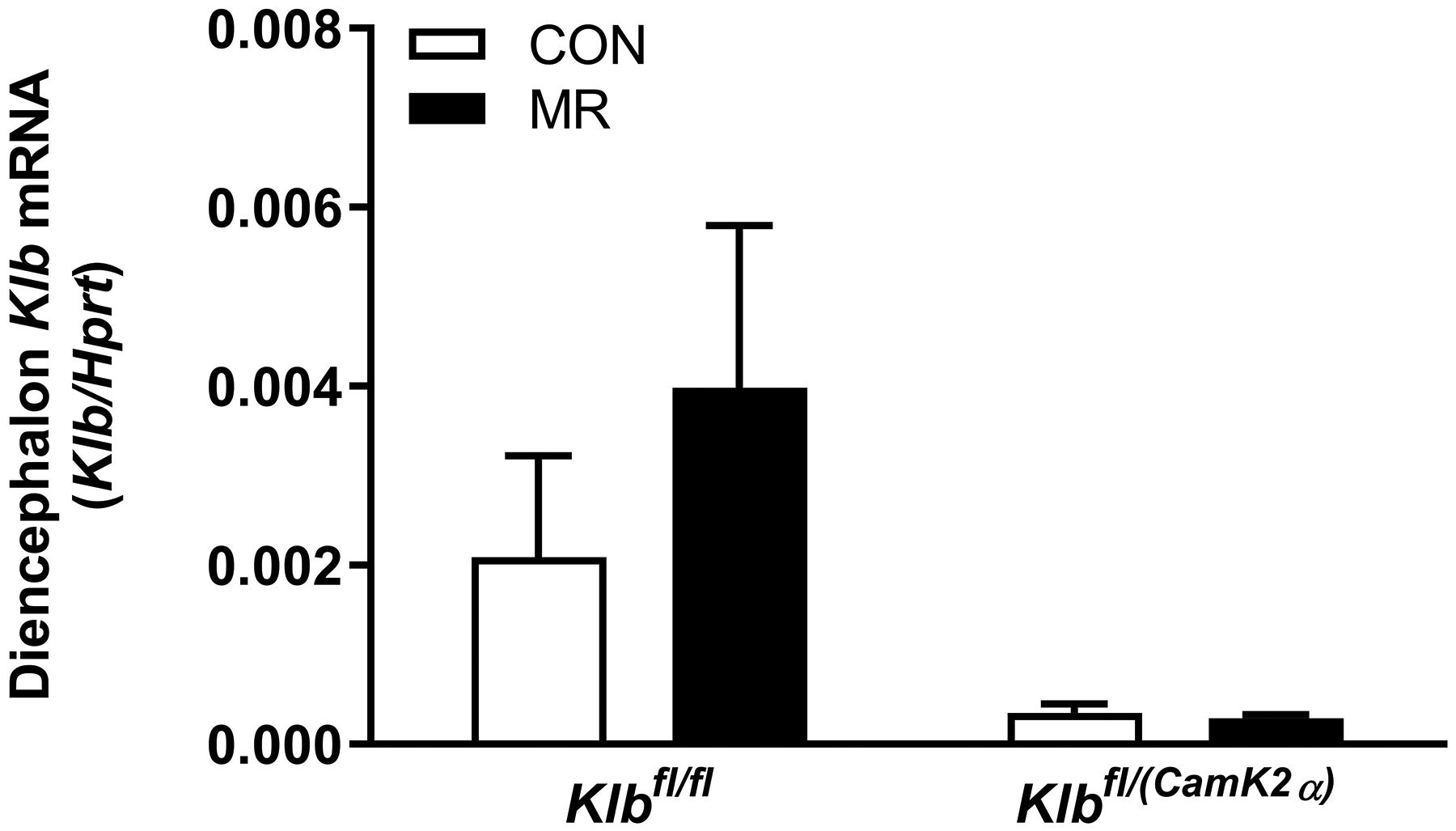
EE was measured via indirect calorimetry (IDC) during the first 12 days of the experiment in sixteen Klbfl/fl and sixteen Klbfl/(Adip) mice on Con and MR diets and for the last 7 days of the experiment in each genotype x diet combination. At the beginning of Experiment 1, Control mice (Klbfl/fl) and adipocyte-specific Klb knockout mice (Klbfl/(Adip)) were placed in the calorimeter chambers for 4 days with all animals receiving the control diet (Con). Then on day 0, half the mice of each genotype were switched to the MR diet while the other half of the mice continued on the Con diet. EE was measured for the next 9 days (Fig. 1A). Error bars are omitted for clarity in Fig. 1A. To better assess the timing of diet-dependent effects on EE after introduction of dietary MR, the mean daytime and nighttime EE during the 3 day run-in period was subtracted from the daytime and nighttime means for each successive 24 h period for each group after introduction of MR, and plotted as diet-dependent changes in daytime and nighttime EE from days 0 through 8. Figs. 1B and 1C show the change over time in daytime mean EE (Fig. 1B) and nighttime mean EE (Fig. 1C) in the Klbfl/fl and Klbfl/(Adip) mice on the Con and MR diets. Mean daytime and nighttime EE in the 4 groups were compared using a repeated meaures two-way ANOVA and means annotated with an asterisk on specific days differ from their corresponding genotype on the Con diet on that day at P<0.05. At the end of Experiments 1 (Fig. 1D) and 2 (Fig. 1E), all mice in each experiment were re-acclimated to the IDC chambers for one week, followed by five days of VCO2 and VO2 measurements for calculation of total energy expenditure (EE). Mean 24 h EE was calculated from the measurements made during the last 5 days of each study and compared by ANCOVA as described in Materials and Methods. All values are expressed as mean ± SEM for 8 mice of each genotype × diet combination. In Figs. 1D and 1E, least square means for 24 h EE not sharing a common letter differ at p < 0.05. In Fig. 1F, the transcript abundance of β-klotho was measured in the diencephlon using hypoxanthine guanine phosphoribosyl transferase (Hrpt) as a denominator and determined to be undetectable.
