FIGURE 3.
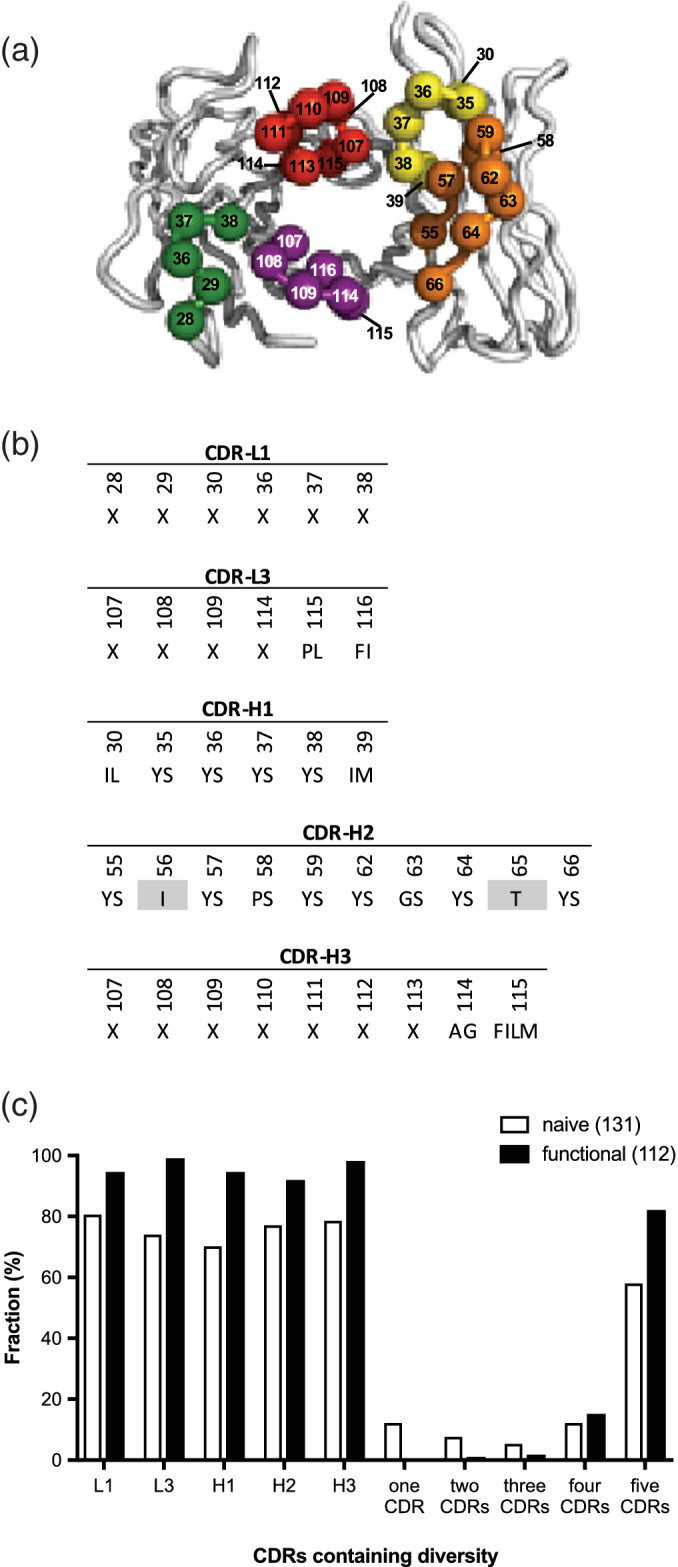
Design and characterization of single‐chain fragment antigen binding (scFab) library R. (a) The backbones of the heavy‐ and light‐chain variable domains are shown as tubes. The framework residues are colored gray and the complement determining region (CDR) loops are colored as follows: CDR‐L1 (green), CDR‐L3 (purple), CDR‐H1 (yellow), CDR‐H2 (orange), and CDR‐H3 (red). Spheres colored according to the CDR coloring scheme represent positions that were diversified. The figure was generated using PyMOL (http://www.pymol.org) with crystal structure coordinates (PDB entry 1FVC). (b) CDR diversity design. Positions shaded in gray were fixed as indicated, and at each diversified position, the allowed amino acids are denoted by the single‐letter code. X denotes a mixture of nine amino acids as follows: Tyr (25%), Ser (20%), Gly (20%), Ala (10%), and Phe, Trp, His, Val, and Pro (5% each). The lengths of CDR‐L1, CDR‐L3, and CDR‐H3 were varied by replacing the positions denoted by X with 5–6, 3–7, or 1–17 degenerate codons, respectively. Residue numbering is according to the IMGT scheme. (c) The fractions of clones containing diversity within a particular CDR or the indicated number of CDRs is shown for 131 naive clones and 112 functional clones that include those shown in Figures 4 and 5
