Fig 4. Frequency distribution of the number of SNPs with near-independent significant effect on stature (P < 5x10-8) in 1,000 control tests for COJO-C genes.
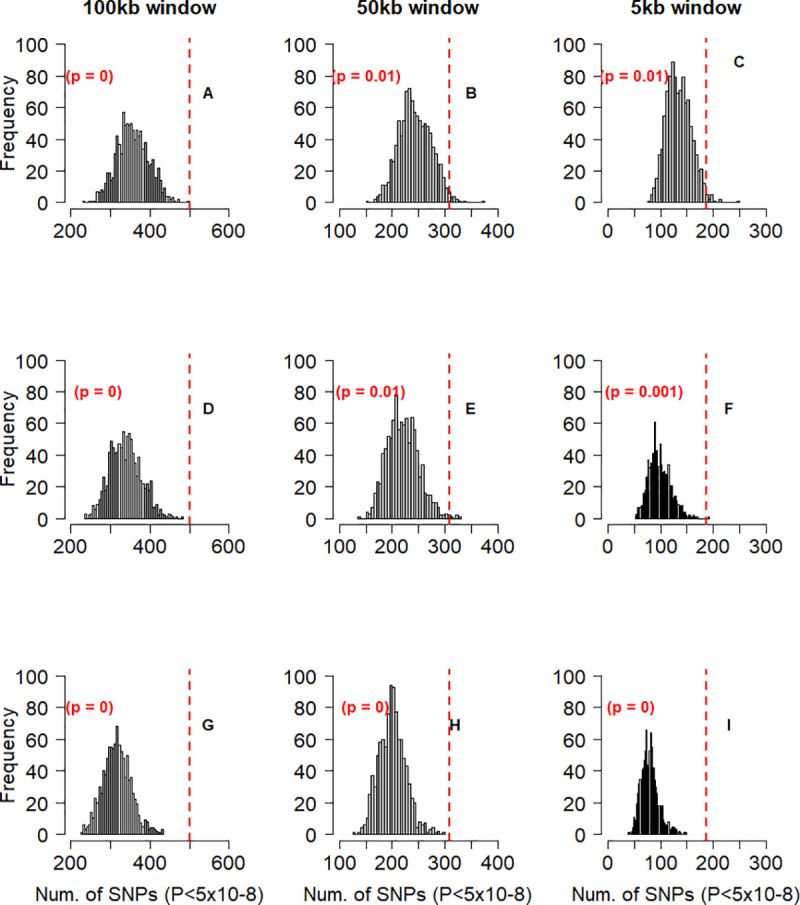
In each test, 1,184 cattle genes were sampled from a total of 13,865 genes that have human orthologs, and near-independent significant SNPs within and 100kbp (Column 1 –panels A,D,G), 50kbp (Column 2 –panels B,E,H) or 5kbp (Column 3 –panels C,F,I) either sides of the sampled genes were identified. the genes were sampled using stratified random sampling, with the genes stratified based on gene-size (Row 1 –panels A,B,C), number of SNPs in a gene (Row 2 –panels D,E,F) or gene LD score (Row 3 –panels G,H,I). The vertical red lines represents the number of near-independent significant SNPs (P < 5x10-8) associated with stature, identified within and around the 1,184 cattle (COJO-C) candidate genes that have been shown to be associated with height in humans. The p value represent the proportion of samples (out of 1,000) that have equal or higher level of enrichment for GWAS signals than the COJO-C genes.
