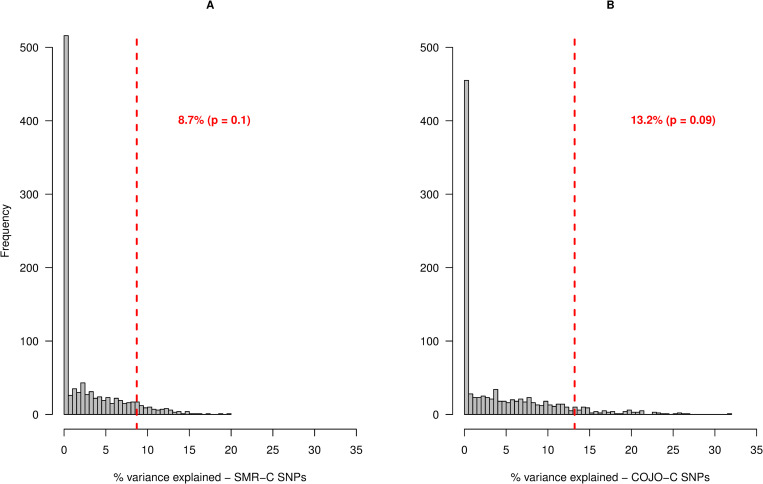
Fig 8. Frequency distribution of the percentage of genetic variance in New Zealand Holstein population, explained by SNPs in 1,000 control tests for COJO-C SNPs.
In each test, SNPs were sampled randomly (matched by allele frequency to candidate SNPs) within a sampled gene-set (5 SNPs were sampled per gene, within each gene-set). SNPs had at least 20 copies of the minor allele in the validation population. Each sampled SNP set was used to construct the first GRM. A second GRM was made with randomly sampled 50k SNPs (excluding all SNPs in the first GRM) across the genome, to account for population structure. Figure A shows the result of control analysis for the SMR-C SNPs, in which 1,777 random SNPs in each gene set were used to construct the first GRM, while figure B shows the result of control analysis for the COJO-C SNPs, in which 5,781 random SNPs within each gene set was used to construct the first GRM. The red vertical lines represents the percentage of genetic variance explained by the candidate SMR-C and COJO-C SNPs, respectively.