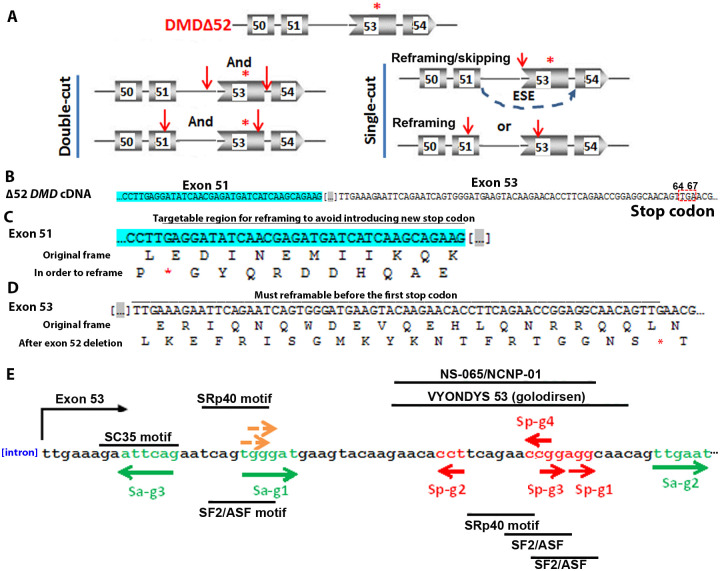
Fig 1. Using CRISPR/Cas9 for restoring dystrophin caused by exon 52 deletion.
A. Single- and double-cut strategies for restoring dystrophin expression after exon 52 deletion. The asterisk in red indicates the first stop codon in the frameshift caused by exon 52 deletion. The red arrows indicate target sites of sgRNAs. Thin lines indicate introns and boxes indicate exons with numbers. “And” indicates two sgRNAs are needed and “or” indicates only one is needed. B. cDNA sequences of exon 51 and exon 53 near the deleted exon 52. The deleted exon 52 sequences are indicated by dots in square brackets. Exon 51 is highlighted green. The first stop codon in exon 53 caused by exon 52 deletion is indicated by a red dashed box. C. Sequence of exon 51 near the deleted exon 52. The original reading frame and the reading frame for reframing in exon 51 are shown. The region below the line is the targetable region that reframing does not introduce new stop codon (red asterisk) in exon 51. D. Sequence of exon 53 near the deleted exon 52. The original reading frame and the reading frame for reframing in exon 53 are shown. The region below the line is the targetable region to make sure that reframing happens before the first stop codon (red asterisk) in exon 53 caused by exon 52 deletion. E. Candidate target sites in exon 53 that can be targeted by Cas9 for reframing dystrophin for exon 52 deletion. PAM regions for SaCa9 and SpCas9 are in green and red respectively. The two brown arrows indicate the PAMs for two possible SpCas9 target sites that have too many predicted off-targets and are not pursued further. The directions of the arrows indicate whether the PAMs are on the sense strand (right directed arrows) or the anti-sense strand (left directed arrows). The target regions for VYONDYS 53 (golodirsen) and NS-065/NCNP-01 are indicated. The predicted ESE motifs near the cleavage sites are listed.